Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
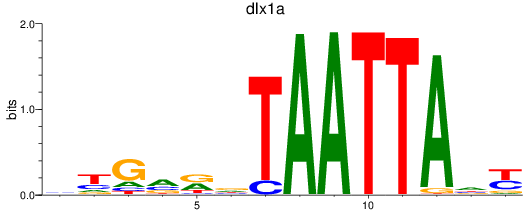
Results for dlx1a
Z-value: 0.28
Transcription factors associated with dlx1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dlx1a
|
ENSDARG00000013125 | distal-less homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dlx1a | dr11_v1_chr9_+_3388099_3388099 | -0.43 | 7.8e-02 | Click! |
Activity profile of dlx1a motif
Sorted Z-values of dlx1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_21001264 | 0.38 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr10_-_21362071 | 0.36 |
ENSDART00000125167
|
avd
|
avidin |
| chr10_-_21362320 | 0.34 |
ENSDART00000189789
|
avd
|
avidin |
| chr9_+_44994214 | 0.26 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr8_-_23780334 | 0.26 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr10_-_8046764 | 0.25 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr14_-_33481428 | 0.24 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr21_+_25777425 | 0.23 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr12_-_33357655 | 0.22 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr3_+_7808459 | 0.20 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr18_-_40708537 | 0.19 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr11_-_44194132 | 0.19 |
ENSDART00000182954
ENSDART00000111271 |
CABZ01080074.1
|
|
| chr5_+_16117871 | 0.18 |
ENSDART00000090657
|
znrf3
|
zinc and ring finger 3 |
| chr18_+_39487486 | 0.18 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr17_+_16046314 | 0.18 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr9_-_712308 | 0.18 |
ENSDART00000144625
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr19_-_18313303 | 0.17 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr4_+_5156117 | 0.17 |
ENSDART00000067392
|
tigarb
|
tp53-induced glycolysis and apoptosis regulator b |
| chr11_-_6452444 | 0.17 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr21_-_32060993 | 0.17 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr23_-_44574059 | 0.17 |
ENSDART00000123007
|
si:ch73-160p18.3
|
si:ch73-160p18.3 |
| chr11_-_1550709 | 0.17 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr2_-_15324837 | 0.16 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr12_-_13729263 | 0.16 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr11_-_23687158 | 0.16 |
ENSDART00000189599
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr4_-_16628801 | 0.16 |
ENSDART00000040708
ENSDART00000064009 |
caprin2
|
caprin family member 2 |
| chr14_-_25935167 | 0.16 |
ENSDART00000139855
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr3_-_40054615 | 0.16 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr17_+_16046132 | 0.16 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr19_-_25149598 | 0.15 |
ENSDART00000162917
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr25_+_36292057 | 0.15 |
ENSDART00000152329
|
bmb
|
brambleberry |
| chr25_+_5972690 | 0.15 |
ENSDART00000067517
|
si:ch211-11i22.4
|
si:ch211-11i22.4 |
| chr11_+_18175893 | 0.15 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr10_-_8053753 | 0.15 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr23_+_36460239 | 0.15 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr10_+_44699734 | 0.15 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr11_-_44801968 | 0.15 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr10_-_8053385 | 0.14 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr10_+_44700103 | 0.14 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr3_-_61494840 | 0.14 |
ENSDART00000101957
|
baiap2l1b
|
BAI1-associated protein 2-like 1b |
| chr6_+_3809346 | 0.14 |
ENSDART00000185105
|
FO704755.1
|
|
| chr7_-_48251234 | 0.14 |
ENSDART00000024062
ENSDART00000098904 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr5_+_44805269 | 0.14 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr10_+_15024772 | 0.14 |
ENSDART00000135667
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr18_+_924949 | 0.14 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr13_+_25397098 | 0.13 |
ENSDART00000132953
|
gsto2
|
glutathione S-transferase omega 2 |
| chr6_+_4528631 | 0.13 |
ENSDART00000122042
|
RNF219
|
ring finger protein 219 |
| chr6_+_19948043 | 0.13 |
ENSDART00000182636
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr10_+_15025006 | 0.13 |
ENSDART00000145192
ENSDART00000140084 |
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr19_-_25149034 | 0.13 |
ENSDART00000148432
ENSDART00000175266 |
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr22_-_22147375 | 0.13 |
ENSDART00000149304
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr13_+_25396896 | 0.13 |
ENSDART00000041257
|
gsto2
|
glutathione S-transferase omega 2 |
| chr2_+_5371492 | 0.13 |
ENSDART00000139762
|
si:ch1073-184j22.1
|
si:ch1073-184j22.1 |
| chr8_+_2642384 | 0.13 |
ENSDART00000143242
|
naif1
|
nuclear apoptosis inducing factor 1 |
| chr7_+_29163762 | 0.12 |
ENSDART00000173762
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr20_-_14114078 | 0.12 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr12_+_23812530 | 0.12 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr12_-_48188928 | 0.12 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr14_-_33945692 | 0.12 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr12_+_23912074 | 0.12 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr18_+_8912710 | 0.12 |
ENSDART00000142866
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr10_+_213878 | 0.12 |
ENSDART00000135903
ENSDART00000138812 |
mpzl1l
|
myelin protein zero-like 1 like |
| chr8_+_32747612 | 0.12 |
ENSDART00000142824
|
hmcn2
|
hemicentin 2 |
| chr16_+_35916371 | 0.12 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
| chr3_-_26183699 | 0.12 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr5_-_7513082 | 0.11 |
ENSDART00000158913
|
bmpr1ba
|
bone morphogenetic protein receptor, type IBa |
| chr20_+_36812368 | 0.11 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr11_-_38533505 | 0.11 |
ENSDART00000113894
|
slc45a3
|
solute carrier family 45, member 3 |
| chr14_-_16082806 | 0.11 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr11_-_6877973 | 0.11 |
ENSDART00000160271
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr10_+_44363195 | 0.11 |
ENSDART00000161744
|
kmt5aa
|
lysine methyltransferase 5Aa |
| chr18_+_18000887 | 0.11 |
ENSDART00000147797
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr9_-_11676491 | 0.11 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr12_+_4920451 | 0.11 |
ENSDART00000171525
ENSDART00000159986 |
plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr5_+_65970235 | 0.11 |
ENSDART00000166432
|
slc2a8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr21_+_43702016 | 0.11 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr16_+_39146696 | 0.11 |
ENSDART00000121756
ENSDART00000084381 |
sybu
|
syntabulin (syntaxin-interacting) |
| chr3_+_7771420 | 0.11 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr22_-_10541712 | 0.11 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr8_+_23826985 | 0.11 |
ENSDART00000187430
|
mapk14a
|
mitogen-activated protein kinase 14a |
| chr8_+_16676894 | 0.11 |
ENSDART00000076586
|
si:ch211-198n5.11
|
si:ch211-198n5.11 |
| chr9_+_54039006 | 0.11 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr14_+_8940326 | 0.10 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr16_-_27677930 | 0.10 |
ENSDART00000145991
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr18_+_3579829 | 0.10 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr6_+_36839509 | 0.10 |
ENSDART00000190605
ENSDART00000104160 |
zgc:110788
|
zgc:110788 |
| chr22_-_22337382 | 0.10 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr7_+_69459759 | 0.10 |
ENSDART00000160500
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr18_+_8912113 | 0.10 |
ENSDART00000147467
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr7_-_8712148 | 0.10 |
ENSDART00000065488
|
tex261
|
testis expressed 261 |
| chr9_-_49964810 | 0.10 |
ENSDART00000167098
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr11_+_5468629 | 0.10 |
ENSDART00000013203
|
cse1l
|
CSE1 chromosome segregation 1-like (yeast) |
| chr4_-_20177868 | 0.10 |
ENSDART00000003621
|
sinup
|
siaz-interacting nuclear protein |
| chr11_+_17984354 | 0.10 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr18_+_8912536 | 0.10 |
ENSDART00000134827
ENSDART00000061904 |
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr6_+_7533601 | 0.10 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr2_+_6253246 | 0.10 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr15_-_2519640 | 0.10 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr9_-_41040492 | 0.10 |
ENSDART00000163164
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr12_+_22580579 | 0.10 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr24_-_6024466 | 0.10 |
ENSDART00000040865
|
pdss1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr8_+_3434146 | 0.10 |
ENSDART00000164426
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr24_-_34680956 | 0.10 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr21_+_27513859 | 0.10 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr2_-_57837838 | 0.10 |
ENSDART00000010699
|
sf3a2
|
splicing factor 3a, subunit 2 |
| chr3_-_34136778 | 0.10 |
ENSDART00000131951
|
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr8_-_45867358 | 0.10 |
ENSDART00000132810
|
adam9
|
ADAM metallopeptidase domain 9 |
| chr11_-_35171768 | 0.10 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr17_-_40956035 | 0.10 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr6_-_33924883 | 0.10 |
ENSDART00000132762
ENSDART00000148142 ENSDART00000142213 |
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr21_+_13383413 | 0.10 |
ENSDART00000151345
|
zgc:113162
|
zgc:113162 |
| chr9_-_8297344 | 0.10 |
ENSDART00000180945
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr17_-_7733037 | 0.10 |
ENSDART00000064657
|
stx11a
|
syntaxin 11a |
| chr24_+_19415124 | 0.10 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr23_+_26026383 | 0.10 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr15_+_34592215 | 0.10 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr15_+_34934568 | 0.09 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr1_-_51710225 | 0.09 |
ENSDART00000057601
ENSDART00000152745 |
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr15_+_29140126 | 0.09 |
ENSDART00000060034
|
zgc:113149
|
zgc:113149 |
| chr7_-_55633475 | 0.09 |
ENSDART00000149478
|
galns
|
galactosamine (N-acetyl)-6-sulfatase |
| chr1_+_27690 | 0.09 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr20_-_29532939 | 0.09 |
ENSDART00000049224
ENSDART00000062377 |
taf1b
|
TATA box binding protein (Tbp)-associated factor, RNA polymerase I, B |
| chr15_+_28175638 | 0.09 |
ENSDART00000037119
|
slc46a1
|
solute carrier family 46 (folate transporter), member 1 |
| chr23_-_44470253 | 0.09 |
ENSDART00000176333
|
si:ch1073-228j22.2
|
si:ch1073-228j22.2 |
| chr6_+_23026714 | 0.09 |
ENSDART00000124948
|
srp68
|
signal recognition particle 68 |
| chr2_+_44972720 | 0.09 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr24_-_38110779 | 0.09 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr9_+_22631672 | 0.09 |
ENSDART00000101770
ENSDART00000126015 ENSDART00000145005 |
etv5a
|
ets variant 5a |
| chr1_+_19764995 | 0.09 |
ENSDART00000138276
|
si:ch211-42i9.8
|
si:ch211-42i9.8 |
| chr11_+_17984167 | 0.09 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr3_-_27868183 | 0.09 |
ENSDART00000185812
|
abat
|
4-aminobutyrate aminotransferase |
| chr23_-_36446307 | 0.09 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr20_-_20270191 | 0.09 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr20_+_27087539 | 0.09 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr10_-_35257458 | 0.09 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr11_+_11303458 | 0.09 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr23_+_2906031 | 0.08 |
ENSDART00000109304
|
c23h20orf24
|
c23h20orf24 homolog (H. sapiens) |
| chr6_-_14040136 | 0.08 |
ENSDART00000065361
ENSDART00000179765 |
etv5b
|
ets variant 5b |
| chr17_+_24618640 | 0.08 |
ENSDART00000092925
|
commd9
|
COMM domain containing 9 |
| chr5_+_9224051 | 0.08 |
ENSDART00000139265
|
si:ch211-12e13.12
|
si:ch211-12e13.12 |
| chr21_-_39024754 | 0.08 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr22_+_737211 | 0.08 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr8_-_38317914 | 0.08 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr10_+_43039947 | 0.08 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr3_+_22035863 | 0.08 |
ENSDART00000177169
|
cdc27
|
cell division cycle 27 |
| chr14_-_47248784 | 0.08 |
ENSDART00000135479
|
fstl5
|
follistatin-like 5 |
| chr6_+_49723289 | 0.08 |
ENSDART00000190452
|
stx16
|
syntaxin 16 |
| chr4_-_5108844 | 0.08 |
ENSDART00000132666
ENSDART00000136096 |
tmem209
|
transmembrane protein 209 |
| chr8_-_44611357 | 0.08 |
ENSDART00000063396
|
bag4
|
BCL2 associated athanogene 4 |
| chr18_+_14619544 | 0.08 |
ENSDART00000010821
|
utp4
|
UTP4, small subunit processome component |
| chr19_-_29887629 | 0.08 |
ENSDART00000066123
|
kpna6
|
karyopherin alpha 6 (importin alpha 7) |
| chr24_-_2450597 | 0.08 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr11_-_44979281 | 0.08 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr17_-_51818659 | 0.08 |
ENSDART00000111389
ENSDART00000157244 |
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr2_+_24374669 | 0.08 |
ENSDART00000133818
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr4_-_5795309 | 0.08 |
ENSDART00000039987
|
pgm3
|
phosphoglucomutase 3 |
| chr25_-_32869794 | 0.08 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr12_-_314899 | 0.08 |
ENSDART00000066579
|
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr22_+_11775269 | 0.08 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr21_+_34088377 | 0.08 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr8_+_50190742 | 0.08 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr1_-_513762 | 0.08 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr23_+_13528053 | 0.08 |
ENSDART00000162217
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr8_-_25034411 | 0.08 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr23_+_32101202 | 0.08 |
ENSDART00000000992
|
zgc:56699
|
zgc:56699 |
| chr1_+_47165842 | 0.08 |
ENSDART00000053152
ENSDART00000167051 |
cbr1
|
carbonyl reductase 1 |
| chr16_+_42667560 | 0.08 |
ENSDART00000023452
|
dpy19l1l
|
dpy-19-like 1, like (H. sapiens) |
| chr1_-_999556 | 0.08 |
ENSDART00000170884
ENSDART00000172235 |
gart
|
phosphoribosylglycinamide formyltransferase |
| chr19_-_425145 | 0.08 |
ENSDART00000164905
|
dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr2_+_22409249 | 0.08 |
ENSDART00000182915
|
zgc:56628
|
zgc:56628 |
| chr19_-_82504 | 0.08 |
ENSDART00000027864
ENSDART00000160560 |
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr10_+_11261576 | 0.08 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr5_+_6672870 | 0.08 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr13_-_9213207 | 0.07 |
ENSDART00000139861
ENSDART00000140524 |
HTRA2 (1 of many)
|
si:dkey-33c12.11 |
| chr1_+_44196236 | 0.07 |
ENSDART00000179560
ENSDART00000166324 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr25_-_6292270 | 0.07 |
ENSDART00000130809
|
wdr61
|
WD repeat domain 61 |
| chr9_-_54001502 | 0.07 |
ENSDART00000085253
|
mid1
|
midline 1 |
| chr17_+_43867889 | 0.07 |
ENSDART00000132673
ENSDART00000167214 |
zgc:66313
|
zgc:66313 |
| chr25_+_3788443 | 0.07 |
ENSDART00000189747
|
chid1
|
chitinase domain containing 1 |
| chr15_-_44601331 | 0.07 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr19_-_2404599 | 0.07 |
ENSDART00000127578
|
twistnb
|
TWIST neighbor |
| chr21_-_4793686 | 0.07 |
ENSDART00000158232
|
notch1a
|
notch 1a |
| chr24_+_42004640 | 0.07 |
ENSDART00000171380
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr7_+_36898622 | 0.07 |
ENSDART00000190773
|
tox3
|
TOX high mobility group box family member 3 |
| chr20_+_38458084 | 0.07 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr12_-_26538823 | 0.07 |
ENSDART00000143213
|
acsf2
|
acyl-CoA synthetase family member 2 |
| chr6_-_55399214 | 0.07 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr6_-_15491579 | 0.07 |
ENSDART00000156439
|
st6gal2b
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2b |
| chr21_+_13387965 | 0.07 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr6_-_15492030 | 0.07 |
ENSDART00000156141
ENSDART00000183992 |
st6gal2b
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2b |
| chr6_+_4370935 | 0.07 |
ENSDART00000192368
|
rbm26
|
RNA binding motif protein 26 |
| chr25_-_27621268 | 0.07 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr1_+_45002971 | 0.07 |
ENSDART00000021336
|
dnaja1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr24_-_25166720 | 0.07 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr14_+_16287968 | 0.07 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr9_-_394088 | 0.07 |
ENSDART00000169014
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr12_+_46582727 | 0.07 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr6_+_37655078 | 0.07 |
ENSDART00000122199
ENSDART00000065127 |
cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr16_-_2390931 | 0.07 |
ENSDART00000149463
|
hace1
|
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| chr4_+_20051478 | 0.07 |
ENSDART00000143642
|
lamtor4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of dlx1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.2 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.1 | 0.2 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.1 | 0.2 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.0 | 0.3 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.2 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.0 | 0.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.0 | 0.0 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.0 | 0.1 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.1 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.1 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.0 | 0.1 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.0 | 0.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 0.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.1 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.1 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.1 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 0.0 | GO:0071047 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.0 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.0 | 0.1 | GO:0007589 | body fluid secretion(GO:0007589) |
| 0.0 | 0.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.1 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.0 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.0 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.0 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.0 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.0 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.0 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.2 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.2 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.0 | 0.0 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.0 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.0 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.3 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.1 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.1 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.1 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0047760 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.0 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.0 | 0.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0015157 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.0 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.0 | 0.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.0 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |