Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
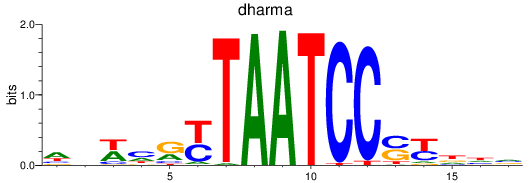
Results for dharma
Z-value: 0.34
Transcription factors associated with dharma
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dharma
|
ENSDARG00000040955 | dharma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dharma | dr11_v1_chr15_-_29012493_29012493 | 0.16 | 5.3e-01 | Click! |
Activity profile of dharma motif
Sorted Z-values of dharma motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_39963599 | 0.41 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr23_+_20705849 | 0.37 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
| chr23_+_44644911 | 0.36 |
ENSDART00000140799
|
zgc:85858
|
zgc:85858 |
| chr5_+_9037650 | 0.35 |
ENSDART00000158226
|
si:ch211-155m12.1
|
si:ch211-155m12.1 |
| chr3_-_14498295 | 0.35 |
ENSDART00000172102
|
elof1
|
ELF1 homolog, elongation factor 1 |
| chr7_+_66884291 | 0.32 |
ENSDART00000187499
|
sbf2
|
SET binding factor 2 |
| chr7_+_66884570 | 0.31 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr10_-_9961488 | 0.29 |
ENSDART00000191023
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr16_+_23403602 | 0.29 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr24_-_24060460 | 0.29 |
ENSDART00000142813
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr24_-_24271629 | 0.27 |
ENSDART00000135060
|
rps6ka3b
|
ribosomal protein S6 kinase, polypeptide 3b |
| chr24_-_24060632 | 0.27 |
ENSDART00000090514
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr8_+_2656231 | 0.23 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr18_-_15551360 | 0.22 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr16_-_21915223 | 0.22 |
ENSDART00000170634
|
setdb1b
|
SET domain, bifurcated 1b |
| chr1_+_292545 | 0.21 |
ENSDART00000148261
|
cenpe
|
centromere protein E |
| chr16_-_22585289 | 0.21 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr2_+_36046126 | 0.21 |
ENSDART00000085976
ENSDART00000171680 |
smg7
|
SMG7 nonsense mediated mRNA decay factor |
| chr20_-_3915770 | 0.21 |
ENSDART00000159322
|
si:ch73-111k22.3
|
si:ch73-111k22.3 |
| chr18_-_7948188 | 0.20 |
ENSDART00000091805
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr3_+_1209006 | 0.20 |
ENSDART00000158633
|
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr18_+_7639401 | 0.20 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr16_+_28547157 | 0.20 |
ENSDART00000109450
ENSDART00000165687 |
fam171a1
|
family with sequence similarity 171, member A1 |
| chr18_+_6706140 | 0.19 |
ENSDART00000111343
|
lmf2a
|
lipase maturation factor 2a |
| chr18_+_50525109 | 0.19 |
ENSDART00000098390
|
ubl7b
|
ubiquitin-like 7b (bone marrow stromal cell-derived) |
| chr6_-_17999776 | 0.19 |
ENSDART00000183048
ENSDART00000181577 ENSDART00000170597 |
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr18_+_6641542 | 0.18 |
ENSDART00000160379
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr5_-_33035295 | 0.18 |
ENSDART00000061153
|
glipr2
|
GLI pathogenesis-related 2 |
| chr16_+_23404170 | 0.17 |
ENSDART00000170061
|
s100w
|
S100 calcium binding protein W |
| chr2_-_32387441 | 0.17 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr9_+_24920677 | 0.17 |
ENSDART00000037025
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr9_-_30502010 | 0.16 |
ENSDART00000149483
|
si:dkey-229b18.3
|
si:dkey-229b18.3 |
| chr14_-_47963115 | 0.16 |
ENSDART00000003826
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr19_+_27859546 | 0.15 |
ENSDART00000161908
|
nsun2
|
NOP2/Sun RNA methyltransferase family, member 2 |
| chr5_-_69716501 | 0.15 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr5_+_13394543 | 0.14 |
ENSDART00000051669
ENSDART00000135921 |
tctn2
|
tectonic family member 2 |
| chr5_-_40040488 | 0.14 |
ENSDART00000157755
|
cds1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr11_+_25157374 | 0.14 |
ENSDART00000019450
|
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr14_-_30490465 | 0.14 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr3_+_16664212 | 0.14 |
ENSDART00000013816
|
zgc:55558
|
zgc:55558 |
| chr11_-_2478374 | 0.14 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr5_-_22602780 | 0.14 |
ENSDART00000011699
|
nono
|
non-POU domain containing, octamer-binding |
| chr14_-_3944017 | 0.13 |
ENSDART00000055817
|
si:ch73-49o8.1
|
si:ch73-49o8.1 |
| chr3_-_56030117 | 0.13 |
ENSDART00000113030
ENSDART00000157764 |
cep112
|
centrosomal protein 112 |
| chr13_+_40815012 | 0.13 |
ENSDART00000016960
|
prkg1a
|
protein kinase, cGMP-dependent, type Ia |
| chr18_-_8579907 | 0.12 |
ENSDART00000147284
|
FRMD4A
|
si:ch211-220f12.1 |
| chr15_-_8191992 | 0.12 |
ENSDART00000155381
ENSDART00000190714 |
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr6_-_10863582 | 0.12 |
ENSDART00000135454
ENSDART00000133893 ENSDART00000014521 ENSDART00000164342 |
lnpk
|
lunapark, ER junction formation factor |
| chr17_+_25426091 | 0.12 |
ENSDART00000149241
ENSDART00000121848 |
srrm1
|
serine/arginine repetitive matrix 1 |
| chr14_-_30490763 | 0.12 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr17_-_33414781 | 0.12 |
ENSDART00000142203
ENSDART00000034638 |
ccdc28a
|
coiled-coil domain containing 28A |
| chr2_+_243778 | 0.12 |
ENSDART00000182262
|
CABZ01085887.1
|
|
| chr7_-_40122139 | 0.11 |
ENSDART00000173982
|
si:ch73-174h16.5
|
si:ch73-174h16.5 |
| chr5_+_4016271 | 0.11 |
ENSDART00000113627
ENSDART00000105832 ENSDART00000121415 |
ggnbp2
|
gametogenetin binding protein 2 |
| chr3_+_14512670 | 0.11 |
ENSDART00000161403
|
rab3db
|
RAB3D, member RAS oncogene family, b |
| chr21_-_21610857 | 0.11 |
ENSDART00000172333
|
diablob
|
diablo, IAP-binding mitochondrial protein b |
| chr24_+_35195963 | 0.11 |
ENSDART00000158801
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr11_+_1602916 | 0.11 |
ENSDART00000184434
ENSDART00000112597 ENSDART00000192165 |
si:dkey-40c23.2
si:dkey-40c23.3
|
si:dkey-40c23.2 si:dkey-40c23.3 |
| chr14_+_34966598 | 0.10 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr16_+_42829735 | 0.10 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr2_+_9560740 | 0.10 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr5_-_22602979 | 0.10 |
ENSDART00000146287
|
nono
|
non-POU domain containing, octamer-binding |
| chr10_+_2841205 | 0.10 |
ENSDART00000131505
ENSDART00000055869 |
ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr7_-_18598661 | 0.09 |
ENSDART00000182109
|
DTX4
|
si:ch211-119e14.2 |
| chr14_+_32838110 | 0.09 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr25_+_36046287 | 0.09 |
ENSDART00000185324
|
rpgrip1l
|
RPGRIP1-like |
| chr7_-_18656069 | 0.09 |
ENSDART00000021559
|
coro1b
|
coronin, actin binding protein, 1B |
| chr25_-_6389713 | 0.09 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr3_-_13509283 | 0.09 |
ENSDART00000166832
|
slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr14_+_7140997 | 0.09 |
ENSDART00000170994
ENSDART00000129898 |
ctsf
|
cathepsin F |
| chr16_+_5898878 | 0.08 |
ENSDART00000180930
|
ulk4
|
unc-51 like kinase 4 |
| chr17_-_17759138 | 0.08 |
ENSDART00000157128
ENSDART00000123845 |
adck1
|
aarF domain containing kinase 1 |
| chr5_-_15264007 | 0.08 |
ENSDART00000180641
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr19_+_2631565 | 0.08 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr10_+_6013076 | 0.08 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr16_+_29586004 | 0.08 |
ENSDART00000149520
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr4_-_11053543 | 0.08 |
ENSDART00000067262
|
mettl25
|
methyltransferase like 25 |
| chr23_+_27352072 | 0.07 |
ENSDART00000144762
|
si:dkey-9p24.5
|
si:dkey-9p24.5 |
| chr11_-_21585176 | 0.07 |
ENSDART00000045391
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr16_+_42830152 | 0.07 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr14_-_25985698 | 0.07 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr3_-_18575868 | 0.07 |
ENSDART00000122968
|
aqp8b
|
aquaporin 8b |
| chr12_-_19185865 | 0.07 |
ENSDART00000153343
|
zc3h7b
|
zinc finger CCCH-type containing 7B |
| chr15_+_36156986 | 0.07 |
ENSDART00000059791
|
sst1.1
|
somatostatin 1, tandem duplicate 1 |
| chr7_-_28058442 | 0.07 |
ENSDART00000173842
|
si:ch211-235p24.2
|
si:ch211-235p24.2 |
| chr1_-_45177373 | 0.07 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr17_+_1632294 | 0.07 |
ENSDART00000191959
|
srp14
|
signal recognition particle 14 |
| chr9_+_55154414 | 0.06 |
ENSDART00000182924
|
ANOS1
|
anosmin 1 |
| chr17_-_12764360 | 0.06 |
ENSDART00000003418
|
brms1la
|
breast cancer metastasis-suppressor 1-like a |
| chr9_-_6502491 | 0.06 |
ENSDART00000102672
|
nck2a
|
NCK adaptor protein 2a |
| chr14_+_29581710 | 0.06 |
ENSDART00000188820
ENSDART00000193874 |
TENM2
|
si:dkey-34l15.2 |
| chr7_+_7019911 | 0.06 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr16_+_11151699 | 0.06 |
ENSDART00000140674
|
cicb
|
capicua transcriptional repressor b |
| chr24_-_1010467 | 0.06 |
ENSDART00000114544
|
cdk13
|
cyclin-dependent kinase 13 |
| chr17_-_6349044 | 0.05 |
ENSDART00000081707
|
oct2
|
organic cation transporter 2 |
| chr13_+_9408101 | 0.05 |
ENSDART00000091502
|
si:ch211-243o19.4
|
si:ch211-243o19.4 |
| chr1_+_27690 | 0.05 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr15_+_15516612 | 0.05 |
ENSDART00000016024
|
traf4a
|
tnf receptor-associated factor 4a |
| chr10_+_10677697 | 0.05 |
ENSDART00000188705
|
fam163b
|
family with sequence similarity 163, member B |
| chr1_-_49950643 | 0.05 |
ENSDART00000138301
|
sgms2
|
sphingomyelin synthase 2 |
| chr10_+_24504464 | 0.05 |
ENSDART00000141387
|
mtus2a
|
microtubule associated tumor suppressor candidate 2a |
| chr15_-_8191841 | 0.05 |
ENSDART00000156663
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr8_-_43574935 | 0.04 |
ENSDART00000051139
|
ncor2
|
nuclear receptor corepressor 2 |
| chr14_+_36521005 | 0.04 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr20_-_31308805 | 0.04 |
ENSDART00000147045
|
hpcal1
|
hippocalcin-like 1 |
| chr8_+_37755099 | 0.04 |
ENSDART00000075708
ENSDART00000140966 |
got1l1
|
glutamic-oxaloacetic transaminase 1 like 1 |
| chr15_+_17848590 | 0.04 |
ENSDART00000168940
|
zgc:113279
|
zgc:113279 |
| chr7_-_41881177 | 0.04 |
ENSDART00000174258
ENSDART00000018972 |
zgc:92818
|
zgc:92818 |
| chr1_-_45146834 | 0.04 |
ENSDART00000144997
|
si:ch211-239f4.6
|
si:ch211-239f4.6 |
| chr7_+_38395197 | 0.04 |
ENSDART00000138669
|
cep89
|
centrosomal protein 89 |
| chr22_+_7462997 | 0.04 |
ENSDART00000106082
|
zgc:112368
|
zgc:112368 |
| chr5_+_57442271 | 0.04 |
ENSDART00000097395
|
prpf4
|
PRP4 pre-mRNA processing factor 4 homolog (yeast) |
| chr23_-_46034609 | 0.04 |
ENSDART00000158712
|
zgc:65873
|
zgc:65873 |
| chr9_+_38967248 | 0.04 |
ENSDART00000112298
|
map2
|
microtubule-associated protein 2 |
| chr10_+_24504292 | 0.04 |
ENSDART00000090059
|
mtus2a
|
microtubule associated tumor suppressor candidate 2a |
| chr22_-_17574511 | 0.03 |
ENSDART00000181496
|
pip5k1ca
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a |
| chr7_+_17953589 | 0.03 |
ENSDART00000174778
ENSDART00000113120 |
taf6l
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr3_+_16762483 | 0.03 |
ENSDART00000132732
|
tmem86b
|
transmembrane protein 86B |
| chr2_-_9744081 | 0.03 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr6_-_39653972 | 0.03 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr10_+_19525839 | 0.03 |
ENSDART00000162912
ENSDART00000158512 |
vsig8a
|
V-set and immunoglobulin domain containing 8a |
| chr12_+_27034760 | 0.03 |
ENSDART00000181170
ENSDART00000153054 |
fbrs
|
fibrosin |
| chr2_+_13050225 | 0.03 |
ENSDART00000182237
|
BX324233.1
|
|
| chr12_+_27034594 | 0.03 |
ENSDART00000111679
|
fbrs
|
fibrosin |
| chr4_-_45043910 | 0.02 |
ENSDART00000164068
ENSDART00000123524 ENSDART00000135010 ENSDART00000191778 |
si:ch211-227e10.6
|
si:ch211-227e10.6 |
| chr24_+_37533728 | 0.02 |
ENSDART00000061203
|
rhot2
|
ras homolog family member T2 |
| chr14_-_970853 | 0.02 |
ENSDART00000130801
|
acsl1b
|
acyl-CoA synthetase long chain family member 1b |
| chr19_+_7567763 | 0.02 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr20_-_31067306 | 0.02 |
ENSDART00000014163
|
fndc1
|
fibronectin type III domain containing 1 |
| chr7_+_8456999 | 0.02 |
ENSDART00000172880
|
jac4
|
jacalin 4 |
| chr9_+_32073606 | 0.02 |
ENSDART00000184170
ENSDART00000180355 ENSDART00000110204 ENSDART00000123278 |
pikfyve
|
phosphoinositide kinase, FYVE finger containing |
| chr14_-_25928899 | 0.02 |
ENSDART00000143518
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr7_+_41146560 | 0.02 |
ENSDART00000143285
ENSDART00000173852 ENSDART00000174003 ENSDART00000038487 ENSDART00000173463 ENSDART00000166448 ENSDART00000052274 |
puf60b
|
poly-U binding splicing factor b |
| chr6_-_58757131 | 0.02 |
ENSDART00000083582
|
soat2
|
sterol O-acyltransferase 2 |
| chr7_+_19903924 | 0.01 |
ENSDART00000159112
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr15_-_15469079 | 0.01 |
ENSDART00000132637
ENSDART00000004220 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr3_+_49521106 | 0.01 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr15_-_6976851 | 0.01 |
ENSDART00000158474
ENSDART00000168943 ENSDART00000169944 |
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr4_+_33818711 | 0.01 |
ENSDART00000186452
|
znf1026
|
zinc finger protein 1026 |
| chr1_+_51479128 | 0.01 |
ENSDART00000028018
|
meis1a
|
Meis homeobox 1 a |
| chr4_-_63558838 | 0.01 |
ENSDART00000191708
ENSDART00000190740 ENSDART00000186476 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr4_-_43862121 | 0.01 |
ENSDART00000101961
ENSDART00000183525 |
si:dkey-261p22.1
|
si:dkey-261p22.1 |
| chr25_-_37305738 | 0.01 |
ENSDART00000154185
|
si:dkey-234i14.15
|
si:dkey-234i14.15 |
| chr23_-_28294763 | 0.01 |
ENSDART00000139537
|
znf385a
|
zinc finger protein 385A |
| chr13_+_24853578 | 0.01 |
ENSDART00000145865
|
si:dkey-24f15.2
|
si:dkey-24f15.2 |
| chr4_-_34480599 | 0.01 |
ENSDART00000159602
ENSDART00000182248 |
si:dkeyp-51g9.4
si:ch211-64i20.3
|
si:dkeyp-51g9.4 si:ch211-64i20.3 |
| chr8_+_46610020 | 0.01 |
ENSDART00000082509
|
tas1r2.2
|
taste receptor, type 1, member 2, tandem duplicate 2 |
| chr1_-_47122058 | 0.01 |
ENSDART00000159925
ENSDART00000101143 ENSDART00000176803 |
mhc1zea
|
major histocompatibility complex class I ZEA |
| chr17_-_20979077 | 0.01 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr4_+_43225728 | 0.01 |
ENSDART00000158639
ENSDART00000141339 ENSDART00000137770 ENSDART00000166019 |
si:ch211-76m11.5
|
si:ch211-76m11.5 |
| chr4_+_46812891 | 0.01 |
ENSDART00000189655
ENSDART00000162329 |
si:ch211-215p11.3
|
si:ch211-215p11.3 |
| chr24_+_2519761 | 0.01 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr3_+_22999849 | 0.01 |
ENSDART00000127239
|
si:ch211-237i5.4
|
si:ch211-237i5.4 |
| chr4_+_45389591 | 0.01 |
ENSDART00000162530
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr10_+_3875716 | 0.01 |
ENSDART00000189268
ENSDART00000180624 |
TTC28
|
tetratricopeptide repeat domain 28 |
| chr22_-_10486477 | 0.01 |
ENSDART00000184366
|
aspn
|
asporin (LRR class 1) |
| chr4_-_71214516 | 0.01 |
ENSDART00000169997
|
si:ch211-205a14.7
|
si:ch211-205a14.7 |
| chr4_-_33763221 | 0.01 |
ENSDART00000132613
ENSDART00000191816 ENSDART00000188473 |
si:ch211-197f20.1
|
si:ch211-197f20.1 |
| chr7_-_34448076 | 0.01 |
ENSDART00000170935
|
nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr6_+_52877147 | 0.01 |
ENSDART00000174370
|
or137-8
|
odorant receptor, family H, subfamily 137, member 8 |
| chr4_+_69712223 | 0.01 |
ENSDART00000163035
ENSDART00000164475 |
si:dkey-238o14.1
zgc:174944
|
si:dkey-238o14.1 zgc:174944 |
| chr4_-_56673431 | 0.01 |
ENSDART00000161464
ENSDART00000191926 |
si:ch211-227p7.5
|
si:ch211-227p7.5 |
| chr13_+_9408445 | 0.01 |
ENSDART00000162415
|
si:ch211-243o19.4
|
si:ch211-243o19.4 |
| chr15_+_21252532 | 0.01 |
ENSDART00000162619
ENSDART00000019636 ENSDART00000144901 ENSDART00000138676 ENSDART00000133821 ENSDART00000146967 ENSDART00000143990 ENSDART00000142070 ENSDART00000132373 |
usf1
|
upstream transcription factor 1 |
| chr17_-_51260476 | 0.01 |
ENSDART00000084348
|
trappc12
|
trafficking protein particle complex 12 |
| chr4_-_76186742 | 0.01 |
ENSDART00000163970
|
si:ch211-106j21.4
|
si:ch211-106j21.4 |
| chr21_+_30084823 | 0.01 |
ENSDART00000154573
|
prob1
|
proline-rich basic protein 1 |
| chr8_-_25814263 | 0.01 |
ENSDART00000143397
|
taf10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr20_+_53441935 | 0.01 |
ENSDART00000175214
|
apobb.2
|
apolipoprotein Bb, tandem duplicate 2 |
| chr4_-_75795899 | 0.01 |
ENSDART00000157925
|
si:dkey-261j11.1
|
si:dkey-261j11.1 |
| chr4_+_40383229 | 0.01 |
ENSDART00000126906
ENSDART00000191198 |
si:ch211-218h8.1
|
si:ch211-218h8.1 |
| chr4_-_71554068 | 0.01 |
ENSDART00000167414
|
si:dkey-27n6.1
|
si:dkey-27n6.1 |
| chr18_+_33685119 | 0.00 |
ENSDART00000027307
ENSDART00000145000 |
si:dkey-47k20.4
|
si:dkey-47k20.4 |
| chr4_-_63559311 | 0.00 |
ENSDART00000164415
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr4_-_38828508 | 0.00 |
ENSDART00000151952
|
si:dkey-59l11.10
|
si:dkey-59l11.10 |
| chr4_+_70341246 | 0.00 |
ENSDART00000123408
|
si:ch211-76m11.3
|
si:ch211-76m11.3 |
| chr4_-_38829098 | 0.00 |
ENSDART00000151932
|
si:dkey-59l11.10
|
si:dkey-59l11.10 |
| chr4_+_35227135 | 0.00 |
ENSDART00000161023
ENSDART00000182403 |
si:ch211-276i6.1
|
si:ch211-276i6.1 |
| chr4_+_45388995 | 0.00 |
ENSDART00000150802
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr22_-_6404807 | 0.00 |
ENSDART00000149399
ENSDART00000063420 |
si:rp71-1i20.2
|
si:rp71-1i20.2 |
| chr4_-_71267777 | 0.00 |
ENSDART00000169664
|
si:dkeyp-51g9.4
|
si:dkeyp-51g9.4 |
| chr4_-_34776147 | 0.00 |
ENSDART00000167498
|
si:dkey-146m20.14
|
si:dkey-146m20.14 |
| chr4_+_51201875 | 0.00 |
ENSDART00000189875
ENSDART00000186722 |
BX005380.1
|
|
| chr23_-_26652273 | 0.00 |
ENSDART00000112124
ENSDART00000111029 |
hspg2
|
heparan sulfate proteoglycan 2 |
| chr4_+_57192681 | 0.00 |
ENSDART00000190438
|
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr4_-_52783184 | 0.00 |
ENSDART00000172283
|
si:dkey-4j21.2
|
si:dkey-4j21.2 |
| chr14_-_7141550 | 0.00 |
ENSDART00000172020
|
trpt1
|
tRNA phosphotransferase 1 |
| chr8_+_2773776 | 0.00 |
ENSDART00000156796
ENSDART00000154529 ENSDART00000155092 |
sh3glb2a
|
SH3-domain GRB2-like endophilin B2a |
| chr4_-_75651505 | 0.00 |
ENSDART00000115143
|
si:dkey-71l4.2
|
si:dkey-71l4.2 |
| chr1_-_44928987 | 0.00 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr4_+_54862025 | 0.00 |
ENSDART00000166721
|
si:dkey-56m15.9
|
si:dkey-56m15.9 |
| chr4_+_65487457 | 0.00 |
ENSDART00000162017
ENSDART00000193241 |
si:dkeyp-5g9.1
|
si:dkeyp-5g9.1 |
| chr4_+_36616118 | 0.00 |
ENSDART00000160058
ENSDART00000170506 |
si:dkey-151g22.1
|
si:dkey-151g22.1 |
| chr8_-_46572298 | 0.00 |
ENSDART00000030470
|
sult1st3
|
sulfotransferase family 1, cytosolic sulfotransferase 3 |
| chr4_-_56103062 | 0.00 |
ENSDART00000165784
ENSDART00000190846 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr4_-_67984478 | 0.00 |
ENSDART00000160623
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr19_+_3140313 | 0.00 |
ENSDART00000125504
|
zgc:86598
|
zgc:86598 |
| chr4_+_76973771 | 0.00 |
ENSDART00000142978
|
si:dkey-240n22.3
|
si:dkey-240n22.3 |
| chr4_-_38539463 | 0.00 |
ENSDART00000158211
|
si:ch211-209n20.3
|
si:ch211-209n20.3 |
| chr4_+_67994238 | 0.00 |
ENSDART00000160968
|
si:ch211-133h13.1
|
si:ch211-133h13.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of dharma
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 0.2 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.0 | 0.2 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.1 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.2 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.2 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.0 | GO:0006531 | aspartate metabolic process(GO:0006531) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.0 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |