Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
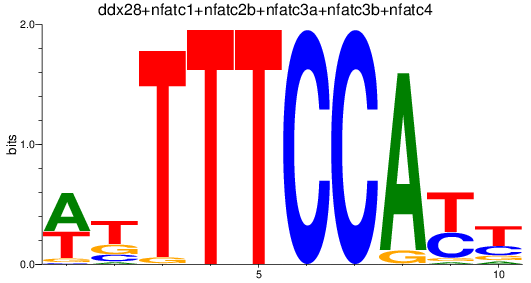
Results for ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4
Z-value: 0.78
Transcription factors associated with ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfatc1
|
ENSDARG00000036168 | nuclear factor of activated T cells 1 |
|
nfatc3b
|
ENSDARG00000051729 | nuclear factor of activated T cells 3b |
|
nfatc4
|
ENSDARG00000054162 | nuclear factor of activated T cells 4 |
|
nfatc3a
|
ENSDARG00000076297 | nuclear factor of activated T cells 3a |
|
nfatc2b
|
ENSDARG00000079972 | nuclear factor of activated T cells 2b |
|
ddx28
|
ENSDARG00000112133 | nuclear factor of activated T cells 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfatc3b | dr11_v1_chr25_-_36248053_36248053 | 0.84 | 1.2e-05 | Click! |
| nfatc2b | dr11_v1_chr6_-_55254786_55254786 | 0.83 | 2.2e-05 | Click! |
| nfatc1 | dr11_v1_chr19_+_22216778_22216778 | 0.74 | 4.3e-04 | Click! |
| nfatc3a | dr11_v1_chr7_-_34927961_34927961 | -0.40 | 9.8e-02 | Click! |
| nfatc4 | dr11_v1_chr2_-_37803614_37803614 | -0.24 | 3.4e-01 | Click! |
Activity profile of ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4 motif
Sorted Z-values of ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_29780752 | 2.03 |
ENSDART00000137400
ENSDART00000145021 |
cfap77
|
cilia and flagella associated protein 77 |
| chr7_-_40110140 | 1.65 |
ENSDART00000173469
|
si:ch73-174h16.5
|
si:ch73-174h16.5 |
| chr24_+_12690163 | 1.59 |
ENSDART00000100644
|
rec8b
|
REC8 meiotic recombination protein b |
| chr15_-_20933574 | 1.50 |
ENSDART00000152648
ENSDART00000152448 ENSDART00000152244 |
usp2a
|
ubiquitin specific peptidase 2a |
| chr5_-_51148298 | 1.49 |
ENSDART00000042420
|
ccdc180
|
coiled-coil domain containing 180 |
| chr7_-_30367650 | 1.32 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr18_-_29961784 | 1.18 |
ENSDART00000135601
ENSDART00000140661 |
si:ch73-103b9.2
|
si:ch73-103b9.2 |
| chr18_-_29962234 | 1.17 |
ENSDART00000144996
|
si:ch73-103b9.2
|
si:ch73-103b9.2 |
| chr6_-_55254786 | 1.09 |
ENSDART00000113805
|
nfatc2b
|
nuclear factor of activated T cells 2b |
| chr19_-_41069573 | 1.06 |
ENSDART00000111982
ENSDART00000193142 |
sgce
|
sarcoglycan, epsilon |
| chr20_+_7209972 | 1.05 |
ENSDART00000189169
ENSDART00000136974 |
si:dkeyp-51f12.3
|
si:dkeyp-51f12.3 |
| chr16_+_23397785 | 1.01 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr16_+_23398369 | 1.00 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr7_-_16597130 | 0.97 |
ENSDART00000144118
|
e2f8
|
E2F transcription factor 8 |
| chr5_+_13353866 | 0.96 |
ENSDART00000099611
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr9_+_26103814 | 0.93 |
ENSDART00000026011
|
efnb2a
|
ephrin-B2a |
| chr12_-_31103187 | 0.91 |
ENSDART00000005562
ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 ENSDART00000122972 ENSDART00000153068 |
tcf7l2
|
transcription factor 7 like 2 |
| chr6_+_49095646 | 0.90 |
ENSDART00000103385
|
slc25a55a
|
solute carrier family 25, member 55a |
| chr4_-_9722568 | 0.89 |
ENSDART00000067190
|
tspan9b
|
tetraspanin 9b |
| chr6_-_39160422 | 0.87 |
ENSDART00000148661
|
stat2
|
signal transducer and activator of transcription 2 |
| chr5_+_20112032 | 0.86 |
ENSDART00000130554
|
isg15
|
ISG15 ubiquitin-like modifier |
| chr12_+_46960579 | 0.86 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr20_-_48516765 | 0.85 |
ENSDART00000150200
|
si:zfos-223e1.2
|
si:zfos-223e1.2 |
| chr16_-_13613475 | 0.84 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr13_+_24717880 | 0.84 |
ENSDART00000147713
|
cfap43
|
cilia and flagella associated protein 43 |
| chr4_+_7677318 | 0.84 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr7_-_16596938 | 0.80 |
ENSDART00000134548
|
e2f8
|
E2F transcription factor 8 |
| chr6_-_39313027 | 0.80 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr6_-_8311044 | 0.79 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr3_-_20091964 | 0.77 |
ENSDART00000029386
ENSDART00000020253 ENSDART00000124326 |
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr11_-_2469529 | 0.77 |
ENSDART00000125677
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr5_-_40910749 | 0.76 |
ENSDART00000083467
ENSDART00000133183 |
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr8_+_25190023 | 0.76 |
ENSDART00000047528
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr3_-_20063671 | 0.75 |
ENSDART00000056614
ENSDART00000126915 |
ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr8_+_18615938 | 0.74 |
ENSDART00000089161
|
si:ch211-232d19.4
|
si:ch211-232d19.4 |
| chr1_-_58592866 | 0.73 |
ENSDART00000161039
|
adgre5b.2
|
adhesion G protein-coupled receptor E5b, duplicate 2 |
| chr7_+_34688527 | 0.73 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr6_+_46258866 | 0.72 |
ENSDART00000134584
|
zgc:162324
|
zgc:162324 |
| chr14_+_24277556 | 0.71 |
ENSDART00000122660
|
hnrnpa0a
|
heterogeneous nuclear ribonucleoprotein A0a |
| chr1_+_44003654 | 0.69 |
ENSDART00000131337
|
si:dkey-22i16.10
|
si:dkey-22i16.10 |
| chr4_+_12013642 | 0.68 |
ENSDART00000067281
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr6_-_31576397 | 0.68 |
ENSDART00000111837
|
raver2
|
ribonucleoprotein, PTB-binding 2 |
| chr22_+_38301365 | 0.66 |
ENSDART00000137339
|
traf5
|
Tnf receptor-associated factor 5 |
| chr16_-_35585374 | 0.65 |
ENSDART00000180208
|
scmh1
|
Scm polycomb group protein homolog 1 |
| chr15_+_36310442 | 0.65 |
ENSDART00000154678
|
gmnc
|
geminin coiled-coil domain containing |
| chr25_+_4837915 | 0.63 |
ENSDART00000168016
|
gnb5a
|
guanine nucleotide binding protein (G protein), beta 5a |
| chr23_+_23119008 | 0.62 |
ENSDART00000132418
|
samd11
|
sterile alpha motif domain containing 11 |
| chr23_-_24682244 | 0.62 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr20_-_5369105 | 0.62 |
ENSDART00000114316
|
sptlc2b
|
serine palmitoyltransferase, long chain base subunit 2b |
| chr1_-_59415948 | 0.61 |
ENSDART00000168614
|
si:ch211-188p14.4
|
si:ch211-188p14.4 |
| chr21_-_43131752 | 0.60 |
ENSDART00000024137
|
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr11_-_29623380 | 0.60 |
ENSDART00000162587
ENSDART00000193935 ENSDART00000191646 |
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr20_-_1176631 | 0.59 |
ENSDART00000152255
|
gabrr2b
|
gamma-aminobutyric acid (GABA) A receptor, rho 2b |
| chr23_-_27633730 | 0.59 |
ENSDART00000103639
|
arf3a
|
ADP-ribosylation factor 3a |
| chr7_-_40993456 | 0.58 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr14_-_33454595 | 0.58 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr3_-_12026741 | 0.57 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr6_-_57539141 | 0.57 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr8_+_7144066 | 0.57 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr1_-_56223913 | 0.56 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr8_+_40628926 | 0.55 |
ENSDART00000163598
|
dusp2
|
dual specificity phosphatase 2 |
| chr4_-_15420452 | 0.53 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr13_+_31497236 | 0.53 |
ENSDART00000146752
|
lrrc9
|
leucine rich repeat containing 9 |
| chr2_-_24369087 | 0.53 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr20_+_38276690 | 0.52 |
ENSDART00000061437
|
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr7_-_52531252 | 0.51 |
ENSDART00000174369
|
tcf12
|
transcription factor 12 |
| chr7_-_39738460 | 0.51 |
ENSDART00000052201
|
ccdc96
|
coiled-coil domain containing 96 |
| chr5_-_68074592 | 0.51 |
ENSDART00000165052
ENSDART00000018792 |
spag7
|
sperm associated antigen 7 |
| chr13_-_11967769 | 0.50 |
ENSDART00000158369
|
ARL3 (1 of many)
|
zgc:110197 |
| chr10_-_28761454 | 0.50 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr11_-_34628789 | 0.49 |
ENSDART00000192433
|
mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr23_-_18017946 | 0.49 |
ENSDART00000104592
|
pm20d1.2
|
peptidase M20 domain containing 1, tandem duplicate 2 |
| chr11_+_13630107 | 0.49 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr6_-_40899618 | 0.49 |
ENSDART00000153949
ENSDART00000021969 |
zgc:172271
|
zgc:172271 |
| chr9_+_3388099 | 0.48 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr25_-_15045338 | 0.48 |
ENSDART00000161165
ENSDART00000165774 ENSDART00000172538 |
pax6a
|
paired box 6a |
| chr19_+_9186175 | 0.48 |
ENSDART00000039325
|
hcn3
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 3 |
| chr11_-_11471857 | 0.48 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr4_-_1497384 | 0.48 |
ENSDART00000093236
|
zmp:0000000711
|
zmp:0000000711 |
| chr9_+_31752628 | 0.48 |
ENSDART00000060054
|
itgbl1
|
integrin, beta-like 1 |
| chr22_-_26353916 | 0.48 |
ENSDART00000077958
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr6_-_21091948 | 0.47 |
ENSDART00000057348
|
inha
|
inhibin, alpha |
| chr3_+_26734162 | 0.47 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr16_-_22713152 | 0.47 |
ENSDART00000140953
ENSDART00000143836 |
si:ch211-105c13.3
|
si:ch211-105c13.3 |
| chr4_-_9764767 | 0.46 |
ENSDART00000164328
ENSDART00000147699 |
mical3b
|
microtubule associated monooxygenase, calponin and LIM domain containing 3b |
| chr11_-_18705303 | 0.46 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr1_-_58664854 | 0.46 |
ENSDART00000109528
|
adgre5b.3
|
adhesion G protein-coupled receptor E5b, duplicate 3 |
| chr12_+_17106117 | 0.45 |
ENSDART00000149990
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr3_-_39696066 | 0.45 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr9_+_21243335 | 0.45 |
ENSDART00000114754
|
si:rp71-68n21.12
|
si:rp71-68n21.12 |
| chr22_+_29027037 | 0.44 |
ENSDART00000170071
|
pimr97
|
Pim proto-oncogene, serine/threonine kinase, related 97 |
| chr5_-_27994679 | 0.44 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr13_-_40401870 | 0.44 |
ENSDART00000128951
|
nkx3.3
|
NK3 homeobox 3 |
| chr23_-_20309505 | 0.43 |
ENSDART00000130856
|
lamb2l
|
laminin, beta 2-like |
| chr13_+_13681681 | 0.43 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr22_-_15587360 | 0.43 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr5_-_52813442 | 0.43 |
ENSDART00000169305
|
zgc:158260
|
zgc:158260 |
| chr24_+_16149514 | 0.43 |
ENSDART00000152087
|
si:dkey-118j18.2
|
si:dkey-118j18.2 |
| chr8_+_53464216 | 0.43 |
ENSDART00000169514
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr1_-_47089818 | 0.41 |
ENSDART00000132378
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr9_-_11263228 | 0.41 |
ENSDART00000113847
|
chpfa
|
chondroitin polymerizing factor a |
| chr24_-_17029374 | 0.41 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr19_-_10243148 | 0.41 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr3_+_45687266 | 0.41 |
ENSDART00000131652
|
gpr146
|
G protein-coupled receptor 146 |
| chr24_+_16149251 | 0.41 |
ENSDART00000188289
|
si:dkey-118j18.2
|
si:dkey-118j18.2 |
| chr8_+_17078692 | 0.41 |
ENSDART00000023206
|
plk2b
|
polo-like kinase 2b (Drosophila) |
| chr25_-_12635371 | 0.40 |
ENSDART00000162463
|
zcchc14
|
zinc finger, CCHC domain containing 14 |
| chr15_-_46821568 | 0.40 |
ENSDART00000148525
|
chrd
|
chordin |
| chr1_-_54425791 | 0.40 |
ENSDART00000039911
|
pkd1a
|
polycystic kidney disease 1a |
| chr13_+_35690023 | 0.40 |
ENSDART00000128865
ENSDART00000130050 |
psme4a
|
proteasome activator subunit 4a |
| chr3_+_52442102 | 0.39 |
ENSDART00000155455
|
si:ch211-191c10.1
|
si:ch211-191c10.1 |
| chr7_-_52558495 | 0.39 |
ENSDART00000138263
ENSDART00000009938 ENSDART00000174292 ENSDART00000174218 ENSDART00000174335 |
tcf12
|
transcription factor 12 |
| chr25_-_35524166 | 0.39 |
ENSDART00000156782
|
slc5a12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr2_+_21486529 | 0.38 |
ENSDART00000047468
|
inhbab
|
inhibin, beta Ab |
| chr19_-_11336782 | 0.38 |
ENSDART00000131014
|
sept7a
|
septin 7a |
| chr14_-_493029 | 0.38 |
ENSDART00000115093
|
zgc:172215
|
zgc:172215 |
| chr8_+_22931427 | 0.38 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr21_+_11401247 | 0.38 |
ENSDART00000143952
|
cel.1
|
carboxyl ester lipase, tandem duplicate 1 |
| chr14_-_4120636 | 0.37 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr19_-_4851411 | 0.37 |
ENSDART00000110398
|
fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr10_+_33990572 | 0.37 |
ENSDART00000138547
|
fryb
|
furry homolog b (Drosophila) |
| chr12_-_5448993 | 0.37 |
ENSDART00000181802
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr5_+_51443009 | 0.37 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr5_+_37091626 | 0.37 |
ENSDART00000161054
|
tagln2
|
transgelin 2 |
| chr7_-_26308098 | 0.37 |
ENSDART00000146440
ENSDART00000146935 ENSDART00000164627 |
zgc:77439
|
zgc:77439 |
| chr4_+_23223881 | 0.36 |
ENSDART00000133056
ENSDART00000089126 |
trhde.1
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 1 |
| chr17_+_28883353 | 0.36 |
ENSDART00000110322
|
prkd1
|
protein kinase D1 |
| chr17_-_6518962 | 0.36 |
ENSDART00000176137
|
CR628323.3
|
|
| chr13_-_8888334 | 0.36 |
ENSDART00000059881
|
plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr10_+_44581378 | 0.35 |
ENSDART00000190331
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr13_+_1155536 | 0.35 |
ENSDART00000148356
|
perp
|
PERP, TP53 apoptosis effector |
| chr22_+_26263290 | 0.35 |
ENSDART00000184840
|
BX649315.1
|
|
| chr7_+_27059330 | 0.35 |
ENSDART00000173919
|
plekha7b
|
pleckstrin homology domain containing, family A member 7b |
| chr13_-_42749916 | 0.35 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr24_-_21819010 | 0.34 |
ENSDART00000091096
|
CR352265.1
|
|
| chr25_-_7759453 | 0.34 |
ENSDART00000142439
ENSDART00000021577 |
phf21ab
|
PHD finger protein 21Ab |
| chr14_+_49296052 | 0.34 |
ENSDART00000006073
ENSDART00000105346 |
anxa6
|
annexin A6 |
| chr21_-_26250443 | 0.34 |
ENSDART00000123713
|
gria1b
|
glutamate receptor, ionotropic, AMPA 1b |
| chr7_+_52761841 | 0.34 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr11_-_37359416 | 0.34 |
ENSDART00000159184
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr3_-_39152478 | 0.34 |
ENSDART00000154550
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr25_+_4812685 | 0.34 |
ENSDART00000193103
|
myo5c
|
myosin VC |
| chr16_-_16225260 | 0.33 |
ENSDART00000165790
|
gra
|
granulito |
| chr5_-_18962794 | 0.33 |
ENSDART00000145210
|
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr19_+_5674907 | 0.33 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr13_+_23157053 | 0.33 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr21_-_17603182 | 0.33 |
ENSDART00000020048
ENSDART00000177270 |
gsna
|
gelsolin a |
| chr16_+_40301056 | 0.33 |
ENSDART00000058578
|
rspo3
|
R-spondin 3 |
| chr7_+_18176162 | 0.32 |
ENSDART00000109171
|
rce1a
|
Ras converting CAAX endopeptidase 1a |
| chr21_+_11883336 | 0.32 |
ENSDART00000151757
|
dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr19_+_19652439 | 0.32 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr17_+_24848976 | 0.32 |
ENSDART00000062917
|
cx35.4
|
connexin 35.4 |
| chr24_+_26887487 | 0.32 |
ENSDART00000189425
|
CR376848.1
|
|
| chr9_-_12424791 | 0.32 |
ENSDART00000135447
ENSDART00000088199 |
zgc:162707
|
zgc:162707 |
| chr12_-_19151708 | 0.31 |
ENSDART00000057124
|
tefa
|
thyrotrophic embryonic factor a |
| chr6_-_50204262 | 0.31 |
ENSDART00000163648
|
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr13_-_46991577 | 0.30 |
ENSDART00000114748
|
vip
|
vasoactive intestinal peptide |
| chr4_+_61995745 | 0.30 |
ENSDART00000171539
|
CT990567.1
|
|
| chr2_-_22927958 | 0.30 |
ENSDART00000141621
|
myo7bb
|
myosin VIIBb |
| chr13_-_31470439 | 0.29 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr25_+_14087045 | 0.29 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr25_+_35683956 | 0.29 |
ENSDART00000149768
|
kif21a
|
kinesin family member 21A |
| chr18_+_41495841 | 0.29 |
ENSDART00000098671
|
si:ch211-203b8.6
|
si:ch211-203b8.6 |
| chr1_-_38816685 | 0.29 |
ENSDART00000075230
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr7_-_13882988 | 0.29 |
ENSDART00000169828
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr13_-_50139916 | 0.28 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr19_+_3056450 | 0.28 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr14_+_26759332 | 0.27 |
ENSDART00000088484
|
ahnak
|
AHNAK nucleoprotein |
| chr9_-_8454060 | 0.27 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr14_-_30587814 | 0.27 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr3_+_39099716 | 0.27 |
ENSDART00000083388
|
tmem98
|
transmembrane protein 98 |
| chr6_-_40885496 | 0.27 |
ENSDART00000189857
|
sirt4
|
sirtuin 4 |
| chr6_-_48094342 | 0.27 |
ENSDART00000137458
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr16_-_30927454 | 0.27 |
ENSDART00000184015
|
slc45a4
|
solute carrier family 45, member 4 |
| chr7_-_38658411 | 0.27 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr1_+_35790082 | 0.26 |
ENSDART00000085051
|
hhip
|
hedgehog interacting protein |
| chr6_-_36552844 | 0.26 |
ENSDART00000023613
|
her6
|
hairy-related 6 |
| chr6_+_10450000 | 0.26 |
ENSDART00000151288
ENSDART00000187431 ENSDART00000192474 ENSDART00000188214 ENSDART00000184766 ENSDART00000190082 |
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr16_-_12787029 | 0.26 |
ENSDART00000139916
|
foxj2
|
forkhead box J2 |
| chr22_+_17433968 | 0.25 |
ENSDART00000139523
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr4_-_22311610 | 0.25 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr16_-_13004166 | 0.25 |
ENSDART00000133735
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr4_-_23643272 | 0.25 |
ENSDART00000112301
ENSDART00000133184 |
trhde.2
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 2 |
| chr17_-_14700889 | 0.25 |
ENSDART00000179975
|
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr20_+_6142433 | 0.25 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr23_+_36130883 | 0.25 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr9_+_13985567 | 0.25 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr2_-_32558795 | 0.24 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr1_-_12278522 | 0.24 |
ENSDART00000142122
ENSDART00000003825 |
cplx2l
|
complexin 2, like |
| chr13_+_1575276 | 0.24 |
ENSDART00000165987
|
DST
|
dystonin |
| chr20_+_15552657 | 0.24 |
ENSDART00000063912
|
jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr18_+_17663898 | 0.24 |
ENSDART00000021213
|
cpne2
|
copine II |
| chr3_-_26921475 | 0.24 |
ENSDART00000130281
|
ciita
|
class II, major histocompatibility complex, transactivator |
| chr12_+_18906939 | 0.24 |
ENSDART00000186074
|
josd1
|
Josephin domain containing 1 |
| chr19_-_38611814 | 0.24 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr22_+_13886821 | 0.24 |
ENSDART00000130585
ENSDART00000105711 |
sh3bp4a
|
SH3-domain binding protein 4a |
| chr5_+_20922100 | 0.23 |
ENSDART00000131838
|
si:dkey-174n20.1
|
si:dkey-174n20.1 |
| chr15_-_34845414 | 0.23 |
ENSDART00000009892
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr17_-_25382367 | 0.23 |
ENSDART00000162306
ENSDART00000165282 |
lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr5_-_48307804 | 0.23 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
Network of associatons between targets according to the STRING database.
First level regulatory network of ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.3 | 1.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.2 | 0.9 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.2 | 0.8 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 0.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 0.5 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.2 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.6 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.2 | 0.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.6 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.5 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.8 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.3 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.4 | GO:0010712 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.1 | 0.9 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.7 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.9 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.1 | 0.3 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.4 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 0.2 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.2 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 1.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.3 | GO:0048242 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.1 | 0.9 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.1 | 0.2 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine metabolic process(GO:0046098) guanine biosynthetic process(GO:0046099) |
| 0.1 | 0.5 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.2 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.1 | 0.9 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.4 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.4 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.1 | 0.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.6 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.5 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.2 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.8 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0002839 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of mast cell activation(GO:0033005) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.0 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 0.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 1.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.4 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.2 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.6 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.3 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 1.0 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.2 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.4 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 0.1 | GO:0002312 | B cell activation involved in immune response(GO:0002312) isotype switching(GO:0045190) |
| 0.0 | 0.2 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.0 | 0.5 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.1 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.0 | 0.2 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 1.0 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.6 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:1903798 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.0 | 0.4 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.2 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.4 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.3 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 0.1 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.0 | 0.1 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.1 | GO:0010759 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) regulation of odontogenesis(GO:0042481) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.0 | 0.0 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.0 | 0.2 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 1.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.0 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.2 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.4 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.4 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 1.6 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 1.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.6 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.4 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.5 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.6 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.4 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.3 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 1.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.0 | GO:0005929 | cilium(GO:0005929) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.2 | 1.3 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.5 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.6 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 2.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.6 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.3 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.3 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.5 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.9 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.2 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.1 | 0.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.5 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.9 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.0 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.6 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.7 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.8 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 1.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.2 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.6 | GO:0016917 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:1901682 | organic acid:sodium symporter activity(GO:0005343) sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.0 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.5 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 0.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 1.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 1.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |