Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
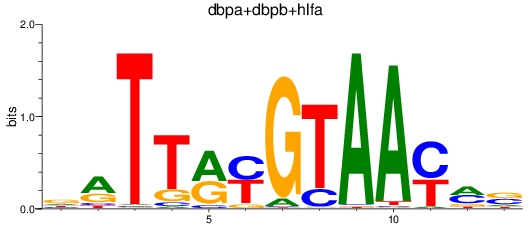
Results for dbpa+dbpb+hlfa
Z-value: 0.73
Transcription factors associated with dbpa+dbpb+hlfa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dbpb
|
ENSDARG00000057652 | D site albumin promoter binding protein b |
|
dbpa
|
ENSDARG00000063014 | D site albumin promoter binding protein a |
|
hlfa
|
ENSDARG00000074752 | HLF transcription factor, PAR bZIP family member a |
|
dbpb
|
ENSDARG00000116703 | D site albumin promoter binding protein b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dbpb | dr11_v1_chr16_-_13612650_13612650 | 0.80 | 7.2e-05 | Click! |
| dbpa | dr11_v1_chr19_-_10196370_10196370 | 0.64 | 4.2e-03 | Click! |
| hlfa | dr11_v1_chr3_-_11624694_11624694 | -0.42 | 8.4e-02 | Click! |
Activity profile of dbpa+dbpb+hlfa motif
Sorted Z-values of dbpa+dbpb+hlfa motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_33361425 | 2.43 |
ENSDART00000031638
|
slc48a1a
|
solute carrier family 48 (heme transporter), member 1a |
| chr16_+_34523515 | 1.35 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr15_+_34988148 | 1.19 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr10_+_506538 | 1.19 |
ENSDART00000141713
|
si:ch211-242f23.3
|
si:ch211-242f23.3 |
| chr9_-_1702648 | 1.13 |
ENSDART00000102934
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr20_+_1121458 | 1.12 |
ENSDART00000064472
|
pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr9_-_9225980 | 1.00 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr18_+_26899316 | 0.99 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr6_+_37754763 | 0.96 |
ENSDART00000110770
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr9_-_48736388 | 0.91 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr14_-_12071679 | 0.83 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr5_-_69212184 | 0.79 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr22_+_10606573 | 0.78 |
ENSDART00000192638
|
rad54l2
|
RAD54 like 2 |
| chr15_-_20939579 | 0.78 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr4_-_12914163 | 0.76 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr17_-_8268406 | 0.74 |
ENSDART00000149873
ENSDART00000064668 ENSDART00000148403 |
ahi1
|
Abelson helper integration site 1 |
| chr19_+_46259619 | 0.71 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr16_+_23397785 | 0.69 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr21_-_32467799 | 0.69 |
ENSDART00000007675
ENSDART00000133099 |
zgc:123105
|
zgc:123105 |
| chr8_+_12930216 | 0.64 |
ENSDART00000115405
|
KIF2A
|
zgc:103670 |
| chr25_+_34014523 | 0.61 |
ENSDART00000182856
|
anxa2a
|
annexin A2a |
| chr22_+_10606863 | 0.59 |
ENSDART00000147975
|
rad54l2
|
RAD54 like 2 |
| chr11_+_39135050 | 0.59 |
ENSDART00000180571
ENSDART00000189685 |
cdc42
|
cell division cycle 42 |
| chr15_-_29348212 | 0.58 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr21_+_43178831 | 0.57 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr5_+_69808763 | 0.56 |
ENSDART00000143482
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr12_-_20373058 | 0.54 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr16_+_23398369 | 0.54 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr6_+_44197099 | 0.50 |
ENSDART00000124168
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr19_+_9032073 | 0.50 |
ENSDART00000127755
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr14_+_4151379 | 0.49 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr6_+_44197348 | 0.48 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr8_+_247163 | 0.48 |
ENSDART00000122378
|
cep120
|
centrosomal protein 120 |
| chr18_-_39288894 | 0.48 |
ENSDART00000186216
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr8_-_53044300 | 0.47 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr6_-_41085692 | 0.45 |
ENSDART00000181463
|
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr5_-_36949476 | 0.45 |
ENSDART00000047269
|
h3f3c
|
H3 histone, family 3C |
| chr23_+_9522781 | 0.44 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr21_-_32467099 | 0.43 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr8_-_41228530 | 0.43 |
ENSDART00000165949
ENSDART00000173055 |
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr8_-_11324143 | 0.42 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr7_+_67733759 | 0.41 |
ENSDART00000172015
|
cyb5b
|
cytochrome b5 type B |
| chr22_+_17531214 | 0.40 |
ENSDART00000161174
ENSDART00000088356 |
hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr24_-_39772045 | 0.39 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr4_+_5341592 | 0.37 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr5_+_22974019 | 0.37 |
ENSDART00000147157
ENSDART00000020434 |
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr23_-_18981595 | 0.37 |
ENSDART00000147617
|
bcl2l1
|
bcl2-like 1 |
| chr16_-_33105677 | 0.36 |
ENSDART00000145055
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr8_-_31716302 | 0.36 |
ENSDART00000061832
|
si:dkey-46a10.3
|
si:dkey-46a10.3 |
| chr1_-_59411901 | 0.36 |
ENSDART00000167015
|
si:ch211-188p14.4
|
si:ch211-188p14.4 |
| chr5_-_15494164 | 0.35 |
ENSDART00000140668
ENSDART00000188076 ENSDART00000085943 |
taok3a
|
TAO kinase 3a |
| chr6_+_48348415 | 0.35 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr23_+_33963619 | 0.35 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr24_+_14581864 | 0.35 |
ENSDART00000134536
|
thtpa
|
thiamine triphosphatase |
| chr1_-_35928942 | 0.32 |
ENSDART00000033566
|
smad1
|
SMAD family member 1 |
| chr10_+_18911152 | 0.32 |
ENSDART00000030205
|
bnip3lb
|
BCL2 interacting protein 3 like b |
| chr22_-_881725 | 0.32 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr13_-_293250 | 0.31 |
ENSDART00000138581
|
chs1
|
chitin synthase 1 |
| chr9_+_1654284 | 0.31 |
ENSDART00000062854
|
nfe2l2a
|
nuclear factor, erythroid 2-like 2a |
| chr16_+_5408748 | 0.31 |
ENSDART00000160008
ENSDART00000014024 |
plecb
|
plectin b |
| chr12_-_26383242 | 0.29 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr7_+_38278860 | 0.29 |
ENSDART00000016265
|
lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr12_-_23365737 | 0.29 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr8_-_53044089 | 0.29 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr19_+_4990320 | 0.28 |
ENSDART00000147056
|
zgc:91968
|
zgc:91968 |
| chr18_-_15551360 | 0.28 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr17_-_17759138 | 0.28 |
ENSDART00000157128
ENSDART00000123845 |
adck1
|
aarF domain containing kinase 1 |
| chr24_-_21343982 | 0.27 |
ENSDART00000012653
|
spice1
|
spindle and centriole associated protein 1 |
| chr12_-_5120175 | 0.27 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr7_+_38380135 | 0.27 |
ENSDART00000174005
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr22_+_15973122 | 0.26 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr16_-_45069882 | 0.26 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr6_+_23810529 | 0.26 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr7_-_7823662 | 0.26 |
ENSDART00000167652
|
cxcl8b.3
|
chemokine (C-X-C motif) ligand 8b, duplicate 3 |
| chr15_+_32711663 | 0.25 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr6_+_23809163 | 0.25 |
ENSDART00000170402
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr17_-_20979077 | 0.25 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr19_+_342094 | 0.25 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr15_+_26600611 | 0.25 |
ENSDART00000155352
|
slc47a3
|
solute carrier family 47 (multidrug and toxin extrusion), member 3 |
| chr1_-_35929143 | 0.25 |
ENSDART00000185002
|
smad1
|
SMAD family member 1 |
| chr17_+_43013171 | 0.24 |
ENSDART00000055541
|
gskip
|
gsk3b interacting protein |
| chr25_-_12803723 | 0.24 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr7_+_69528850 | 0.24 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr15_+_21882419 | 0.23 |
ENSDART00000157216
|
si:dkey-103g5.4
|
si:dkey-103g5.4 |
| chr23_+_16814968 | 0.23 |
ENSDART00000161299
|
zgc:100832
|
zgc:100832 |
| chr3_-_32927516 | 0.23 |
ENSDART00000140117
|
aoc2
|
amine oxidase, copper containing 2 |
| chr17_+_25833947 | 0.22 |
ENSDART00000044328
ENSDART00000154604 |
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr20_-_33675676 | 0.22 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr6_+_9130989 | 0.22 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr13_+_30035253 | 0.22 |
ENSDART00000181303
ENSDART00000057525 ENSDART00000136622 |
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr10_-_24371312 | 0.21 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr20_-_45661049 | 0.21 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr3_-_28665291 | 0.21 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr1_-_10071422 | 0.21 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr22_-_13165186 | 0.21 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr4_+_21867522 | 0.20 |
ENSDART00000140400
|
acss3
|
acyl-CoA synthetase short chain family member 3 |
| chr2_+_10134345 | 0.20 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr6_+_41255485 | 0.20 |
ENSDART00000042683
ENSDART00000186013 |
cadpsb
|
Ca2+-dependent activator protein for secretion b |
| chr3_-_21348478 | 0.20 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr25_-_1235457 | 0.20 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr4_-_12725513 | 0.20 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr5_+_26121393 | 0.19 |
ENSDART00000002221
|
bco2l
|
beta-carotene 15, 15-dioxygenase 2, like |
| chr6_-_14292307 | 0.19 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr11_-_10770053 | 0.19 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr17_-_52091999 | 0.19 |
ENSDART00000019766
|
tgfb3
|
transforming growth factor, beta 3 |
| chr6_+_23809501 | 0.19 |
ENSDART00000168701
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr10_-_25591194 | 0.18 |
ENSDART00000131640
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr25_+_28893615 | 0.18 |
ENSDART00000156994
ENSDART00000075151 |
amn1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr8_+_7801060 | 0.18 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr14_+_21699129 | 0.18 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr9_-_9732212 | 0.18 |
ENSDART00000092884
|
lrrc58b
|
leucine rich repeat containing 58b |
| chr20_-_20610812 | 0.17 |
ENSDART00000181870
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr14_+_7377552 | 0.17 |
ENSDART00000142158
ENSDART00000141471 |
hars
|
histidyl-tRNA synthetase |
| chr22_+_20720808 | 0.17 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr3_-_336299 | 0.17 |
ENSDART00000105021
|
mhc1zfa
|
major histocompatibility complex class I ZFA |
| chr18_+_507618 | 0.17 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr6_+_16468776 | 0.17 |
ENSDART00000109151
ENSDART00000114667 |
zgc:161969
|
zgc:161969 |
| chr16_-_35427060 | 0.17 |
ENSDART00000172294
|
ctps1b
|
CTP synthase 1b |
| chr17_+_24036791 | 0.17 |
ENSDART00000140767
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr11_-_40147032 | 0.17 |
ENSDART00000052918
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr8_+_17884569 | 0.17 |
ENSDART00000134660
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr6_-_7726849 | 0.16 |
ENSDART00000151511
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr23_+_33947874 | 0.16 |
ENSDART00000136104
|
si:ch211-148l7.4
|
si:ch211-148l7.4 |
| chr6_+_10333920 | 0.16 |
ENSDART00000151667
ENSDART00000151477 |
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr8_-_29851706 | 0.16 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr9_-_35069645 | 0.15 |
ENSDART00000122679
ENSDART00000077908 ENSDART00000077894 ENSDART00000125536 |
appb
|
amyloid beta (A4) precursor protein b |
| chr16_+_40954481 | 0.15 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr3_-_36209936 | 0.15 |
ENSDART00000175208
|
csnk1da
|
casein kinase 1, delta a |
| chr5_-_29488245 | 0.15 |
ENSDART00000047719
ENSDART00000141154 ENSDART00000171165 |
cacna1ba
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, a |
| chr21_-_39058490 | 0.15 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr12_-_22400999 | 0.15 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr11_-_45138857 | 0.14 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr25_-_2081371 | 0.14 |
ENSDART00000104915
ENSDART00000156925 |
wnt7bb
|
wingless-type MMTV integration site family, member 7Bb |
| chr3_-_31804481 | 0.14 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr18_+_24562188 | 0.14 |
ENSDART00000099463
|
lysmd4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr7_+_52761841 | 0.14 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr14_-_30905288 | 0.14 |
ENSDART00000173449
ENSDART00000173451 |
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr21_-_39566854 | 0.13 |
ENSDART00000020174
|
dynll2b
|
dynein, light chain, LC8-type 2b |
| chr14_+_21699414 | 0.13 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr25_+_24616717 | 0.13 |
ENSDART00000089113
|
abtb2b
|
ankyrin repeat and BTB (POZ) domain containing 2b |
| chr7_+_19835569 | 0.13 |
ENSDART00000149812
|
ovol1a
|
ovo-like zinc finger 1a |
| chr19_+_7735157 | 0.13 |
ENSDART00000186717
|
tuft1b
|
tuftelin 1b |
| chr25_+_15354095 | 0.13 |
ENSDART00000090397
|
kiaa1549la
|
KIAA1549-like a |
| chr5_+_27583117 | 0.13 |
ENSDART00000180340
|
zmat4a
|
zinc finger, matrin-type 4a |
| chr20_-_20611063 | 0.13 |
ENSDART00000063492
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr7_+_56577906 | 0.12 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr23_-_29812667 | 0.12 |
ENSDART00000006120
|
pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr7_-_41420226 | 0.12 |
ENSDART00000074155
|
abcf2b
|
ATP-binding cassette, sub-family F (GCN20), member 2b |
| chr15_-_22074315 | 0.12 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr11_+_43419809 | 0.12 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr12_+_15002757 | 0.12 |
ENSDART00000135036
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr16_-_7828838 | 0.12 |
ENSDART00000191434
ENSDART00000108653 |
tcaim
|
T cell activation inhibitor, mitochondrial |
| chr8_+_1187928 | 0.12 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr7_-_38792543 | 0.11 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr9_-_13871935 | 0.11 |
ENSDART00000146597
|
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr9_-_6927587 | 0.11 |
ENSDART00000059092
|
tmem182a
|
transmembrane protein 182a |
| chr12_-_9538578 | 0.10 |
ENSDART00000066445
|
nudt13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr2_-_51772438 | 0.10 |
ENSDART00000170241
|
BX908782.2
|
Danio rerio three-finger protein 5 (LOC100003647), mRNA. |
| chr20_-_25533739 | 0.10 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr3_+_19687217 | 0.09 |
ENSDART00000141937
|
tlk2
|
tousled-like kinase 2 |
| chr14_+_16345003 | 0.09 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr3_-_31875138 | 0.09 |
ENSDART00000155298
|
limd2
|
LIM domain containing 2 |
| chr16_-_20294236 | 0.08 |
ENSDART00000059623
|
plekha8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr20_+_25563105 | 0.08 |
ENSDART00000063100
|
cyp2p6
|
cytochrome P450, family 2, subfamily P, polypeptide 6 |
| chr20_-_35246150 | 0.08 |
ENSDART00000090549
|
fzd3a
|
frizzled class receptor 3a |
| chr19_+_41080240 | 0.08 |
ENSDART00000087295
|
ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr12_+_8003872 | 0.08 |
ENSDART00000180300
ENSDART00000126661 |
rhobtb1
|
Rho-related BTB domain containing 1 |
| chr25_-_16826219 | 0.08 |
ENSDART00000191299
ENSDART00000188504 |
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr23_-_30960506 | 0.07 |
ENSDART00000142661
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr20_-_40755614 | 0.07 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr9_+_32301456 | 0.07 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr4_+_21866851 | 0.07 |
ENSDART00000177640
|
acss3
|
acyl-CoA synthetase short chain family member 3 |
| chr16_+_46111849 | 0.07 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr3_-_26184018 | 0.07 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr1_+_2128970 | 0.07 |
ENSDART00000180074
ENSDART00000022019 ENSDART00000098059 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr14_-_9199968 | 0.07 |
ENSDART00000146113
|
arhgef9b
|
Cdc42 guanine nucleotide exchange factor (GEF) 9b |
| chr6_+_22025806 | 0.06 |
ENSDART00000151240
|
ZXDC
|
si:dkey-156n14.3 |
| chr7_+_46020508 | 0.06 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr6_-_40583253 | 0.06 |
ENSDART00000032603
|
tspo
|
translocator protein |
| chr13_+_23157053 | 0.06 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr20_-_1151265 | 0.06 |
ENSDART00000012376
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr2_+_49860722 | 0.06 |
ENSDART00000144060
|
rpl37
|
ribosomal protein L37 |
| chr1_+_31112436 | 0.05 |
ENSDART00000075340
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr4_-_20181964 | 0.05 |
ENSDART00000022539
|
fgl2a
|
fibrinogen-like 2a |
| chr16_+_14029283 | 0.05 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr2_+_36616830 | 0.05 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr4_-_5302866 | 0.05 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr22_-_10121880 | 0.05 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr5_+_3501859 | 0.05 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr23_-_37291793 | 0.05 |
ENSDART00000083281
ENSDART00000187108 |
mul1b
|
mitochondrial E3 ubiquitin protein ligase 1b |
| chr10_+_9595575 | 0.05 |
ENSDART00000091780
ENSDART00000184287 |
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr10_+_10210455 | 0.05 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr25_+_32473277 | 0.05 |
ENSDART00000146451
|
sqor
|
sulfide quinone oxidoreductase |
| chr22_+_15323930 | 0.05 |
ENSDART00000142416
|
si:dkey-236e20.3
|
si:dkey-236e20.3 |
| chr11_+_43751263 | 0.05 |
ENSDART00000163843
|
zgc:153431
|
zgc:153431 |
| chr25_-_11088839 | 0.05 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr23_+_16815353 | 0.04 |
ENSDART00000142789
ENSDART00000143424 |
zgc:100832
|
zgc:100832 |
| chr24_+_18948665 | 0.04 |
ENSDART00000106186
|
prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr1_-_45215343 | 0.04 |
ENSDART00000014727
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr21_-_20733615 | 0.04 |
ENSDART00000145544
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of dbpa+dbpb+hlfa
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.2 | 2.4 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.2 | 0.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.6 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.1 | 0.5 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 0.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 0.7 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.2 | GO:0042364 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 0.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.6 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.2 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.1 | 0.3 | GO:1903523 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.1 | 0.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.3 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.3 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.1 | 0.3 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 0.2 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.1 | 0.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.3 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.3 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 0.3 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.4 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.4 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.3 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:0022602 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.0 | 0.1 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.0 | 0.3 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.2 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.1 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.8 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.0 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.8 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.2 | 0.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.5 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.3 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 2.6 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.0 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.0 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.2 | 0.8 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.8 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.4 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 0.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.7 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.3 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.1 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.3 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.3 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.6 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 1.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.1 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.2 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.3 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.0 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 1.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |