Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
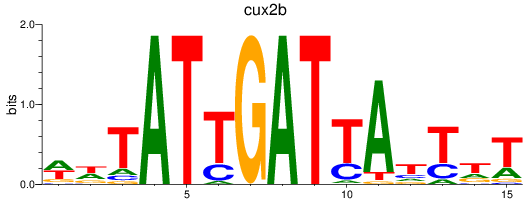
Results for cux2b
Z-value: 1.04
Transcription factors associated with cux2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cux2b
|
ENSDARG00000086345 | cut-like homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cux2b | dr11_v1_chr8_-_4327473_4327473 | -0.44 | 7.0e-02 | Click! |
Activity profile of cux2b motif
Sorted Z-values of cux2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_60721342 | 4.44 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr8_+_21376290 | 4.00 |
ENSDART00000136765
|
ela2
|
elastase 2 |
| chr2_-_38000276 | 3.12 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr14_-_34512859 | 2.89 |
ENSDART00000140368
|
si:ch211-232m8.3
|
si:ch211-232m8.3 |
| chr1_-_54158902 | 2.69 |
ENSDART00000193267
|
CABZ01067151.1
|
|
| chr13_+_50375800 | 2.59 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr8_-_21372446 | 2.43 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr19_+_34274504 | 2.38 |
ENSDART00000132046
|
si:ch211-9n13.3
|
si:ch211-9n13.3 |
| chr25_-_5119162 | 2.29 |
ENSDART00000153961
|
shisal1b
|
shisa like 1b |
| chr22_+_110158 | 2.14 |
ENSDART00000143698
|
prkar2ab
|
protein kinase, cAMP-dependent, regulatory, type II, alpha, B |
| chr16_-_54471235 | 2.10 |
ENSDART00000061572
|
cyp11c1
|
cytochrome P450, family 11, subfamily C, polypeptide 1 |
| chr2_-_42958619 | 2.04 |
ENSDART00000144317
|
oc90
|
otoconin 90 |
| chr18_+_7611298 | 1.96 |
ENSDART00000062156
|
odf3b
|
outer dense fiber of sperm tails 3B |
| chr10_+_2715548 | 1.94 |
ENSDART00000130793
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr9_+_13120419 | 1.92 |
ENSDART00000141005
|
fam117bb
|
family with sequence similarity 117, member Bb |
| chr7_-_4461104 | 1.87 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr4_+_77920666 | 1.85 |
ENSDART00000129523
|
ttll1
|
tubulin tyrosine ligase-like family, member 1 |
| chr12_-_48970299 | 1.77 |
ENSDART00000163734
|
rgrb
|
retinal G protein coupled receptor b |
| chr11_-_22605981 | 1.77 |
ENSDART00000186923
|
myog
|
myogenin |
| chr13_+_7479312 | 1.75 |
ENSDART00000183944
|
cngk
|
cyclic nucleotide-gated potassium channel |
| chr5_-_71705191 | 1.75 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr17_-_681142 | 1.74 |
ENSDART00000165583
|
soul3
|
heme-binding protein soul3 |
| chr11_+_25259058 | 1.71 |
ENSDART00000154109
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr15_-_20933574 | 1.69 |
ENSDART00000152648
ENSDART00000152448 ENSDART00000152244 |
usp2a
|
ubiquitin specific peptidase 2a |
| chr12_+_20149305 | 1.68 |
ENSDART00000126676
ENSDART00000153327 |
foxj1b
|
forkhead box J1b |
| chr21_+_18877130 | 1.65 |
ENSDART00000136893
|
si:dkey-65l23.2
|
si:dkey-65l23.2 |
| chr2_+_2737422 | 1.61 |
ENSDART00000032459
|
aqp1a.1
|
aquaporin 1a (Colton blood group), tandem duplicate 1 |
| chr23_+_45512825 | 1.61 |
ENSDART00000064846
|
prelid1b
|
PRELI domain containing 1b |
| chr19_-_3123963 | 1.59 |
ENSDART00000122816
|
si:ch211-80h18.1
|
si:ch211-80h18.1 |
| chr5_+_35744623 | 1.57 |
ENSDART00000148213
ENSDART00000076627 |
yipf6
|
Yip1 domain family, member 6 |
| chr15_+_21672281 | 1.55 |
ENSDART00000153923
|
si:dkey-40g16.5
|
si:dkey-40g16.5 |
| chr3_-_31783737 | 1.54 |
ENSDART00000090809
|
kcnh6a
|
potassium voltage-gated channel, subfamily H (eag-related), member 6a |
| chr19_+_11978209 | 1.50 |
ENSDART00000111568
|
polr2k
|
polymerase (RNA) II (DNA directed) polypeptide K |
| chr9_+_21194445 | 1.40 |
ENSDART00000061321
|
hsd3b1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr16_-_54942532 | 1.39 |
ENSDART00000078887
ENSDART00000101402 |
tmem222a
|
transmembrane protein 222a |
| chr7_-_34505985 | 1.38 |
ENSDART00000173806
ENSDART00000024869 |
tex9
|
testis expressed 9 |
| chr6_+_515181 | 1.34 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr11_-_1509773 | 1.33 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr3_+_31945604 | 1.32 |
ENSDART00000114957
|
zgc:193811
|
zgc:193811 |
| chr12_+_18533198 | 1.31 |
ENSDART00000189729
|
meiob
|
meiosis specific with OB domains |
| chr6_+_52235441 | 1.31 |
ENSDART00000056319
|
cox6c
|
cytochrome c oxidase subunit VIc |
| chr2_+_44545912 | 1.29 |
ENSDART00000155362
|
si:dkeyp-94h10.5
|
si:dkeyp-94h10.5 |
| chr25_-_17494364 | 1.29 |
ENSDART00000154134
|
lrrc29
|
leucine rich repeat containing 29 |
| chr6_+_13787855 | 1.29 |
ENSDART00000182899
|
tmem198b
|
transmembrane protein 198b |
| chr3_-_34624745 | 1.28 |
ENSDART00000151091
|
tac4
|
tachykinin 4 (hemokinin) |
| chr24_-_21952164 | 1.27 |
ENSDART00000058948
|
acot9.2
|
acyl-CoA thioesterase 9, tandem duplicate 2 |
| chr2_-_44183613 | 1.26 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr21_+_18292535 | 1.26 |
ENSDART00000170205
ENSDART00000169676 ENSDART00000163063 |
dnai1.1
|
dynein, axonemal, intermediate chain 1, paralog 1 |
| chr18_-_20869175 | 1.26 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr21_+_35215810 | 1.25 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr5_-_10946232 | 1.25 |
ENSDART00000163139
ENSDART00000031265 |
rtn4r
|
reticulon 4 receptor |
| chr15_+_21672700 | 1.22 |
ENSDART00000187043
|
si:dkey-40g16.5
|
si:dkey-40g16.5 |
| chr8_-_43120844 | 1.21 |
ENSDART00000193242
|
ccdc92
|
coiled-coil domain containing 92 |
| chr17_+_27545183 | 1.21 |
ENSDART00000129392
|
pacrg
|
PARK2 co-regulated |
| chr6_+_46259950 | 1.19 |
ENSDART00000032326
|
zgc:162324
|
zgc:162324 |
| chr17_-_26610814 | 1.18 |
ENSDART00000133402
ENSDART00000016608 |
mrpl57
|
mitochondrial ribosomal protein L57 |
| chr2_-_44183451 | 1.18 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr20_-_7080427 | 1.16 |
ENSDART00000140166
ENSDART00000023677 |
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr1_-_52498146 | 1.15 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr16_+_54780544 | 1.14 |
ENSDART00000126646
|
si:zfos-1192g2.3
|
si:zfos-1192g2.3 |
| chr20_+_36628059 | 1.14 |
ENSDART00000062898
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr2_-_16565690 | 1.13 |
ENSDART00000022549
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr7_+_29012033 | 1.13 |
ENSDART00000173909
ENSDART00000145762 |
dnaaf1
|
dynein, axonemal, assembly factor 1 |
| chr10_-_25852517 | 1.12 |
ENSDART00000191551
|
trpc4a
|
transient receptor potential cation channel, subfamily C, member 4a |
| chr19_-_48391415 | 1.12 |
ENSDART00000170726
ENSDART00000169577 |
si:ch73-359m17.2
|
si:ch73-359m17.2 |
| chr19_-_42391383 | 1.12 |
ENSDART00000110075
ENSDART00000087002 |
plekho1a
|
pleckstrin homology domain containing, family O member 1a |
| chr7_+_46252993 | 1.12 |
ENSDART00000167149
|
znf536
|
zinc finger protein 536 |
| chr16_-_2650341 | 1.11 |
ENSDART00000128169
ENSDART00000155432 ENSDART00000103722 |
lyplal1
|
lysophospholipase-like 1 |
| chr19_+_48102560 | 1.10 |
ENSDART00000164464
|
utp18
|
UTP18 small subunit (SSU) processome component |
| chr7_-_16598212 | 1.10 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr16_+_50953842 | 1.09 |
ENSDART00000174709
|
si:dkeyp-97a10.1
|
si:dkeyp-97a10.1 |
| chr14_-_52480661 | 1.07 |
ENSDART00000158353
|
exosc3
|
exosome component 3 |
| chr7_-_16597130 | 1.06 |
ENSDART00000144118
|
e2f8
|
E2F transcription factor 8 |
| chr1_+_16073887 | 1.06 |
ENSDART00000160270
|
tusc3
|
tumor suppressor candidate 3 |
| chr2_+_21634128 | 1.05 |
ENSDART00000089822
|
fbxl7
|
F-box and leucine-rich repeat protein 7 |
| chr2_-_45510699 | 1.05 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr17_-_50248023 | 1.05 |
ENSDART00000150024
|
acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr13_-_24328794 | 1.05 |
ENSDART00000145351
|
si:ch211-202m22.1
|
si:ch211-202m22.1 |
| chr19_+_627899 | 1.05 |
ENSDART00000148508
|
tert
|
telomerase reverse transcriptase |
| chr17_+_31592191 | 1.05 |
ENSDART00000153765
|
si:dkey-13p1.3
|
si:dkey-13p1.3 |
| chr9_+_32073606 | 1.04 |
ENSDART00000184170
ENSDART00000180355 ENSDART00000110204 ENSDART00000123278 |
pikfyve
|
phosphoinositide kinase, FYVE finger containing |
| chr10_+_453619 | 1.04 |
ENSDART00000135598
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr10_-_641609 | 1.04 |
ENSDART00000041236
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr13_-_4333181 | 1.04 |
ENSDART00000122406
|
znf318
|
zinc finger protein 318 |
| chr23_+_12134839 | 1.04 |
ENSDART00000128551
ENSDART00000141204 |
ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr14_+_3944826 | 1.04 |
ENSDART00000170167
|
lrp2bp
|
LRP2 binding protein |
| chr2_+_32843133 | 1.03 |
ENSDART00000018501
|
opn4.1
|
opsin 4.1 |
| chr7_+_38395197 | 1.01 |
ENSDART00000138669
|
cep89
|
centrosomal protein 89 |
| chr1_-_47071979 | 1.01 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr16_+_25316973 | 1.01 |
ENSDART00000086409
|
dync1i1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr6_+_26314464 | 0.98 |
ENSDART00000115392
|
dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr12_+_2522642 | 0.98 |
ENSDART00000152567
|
frmpd2
|
FERM and PDZ domain containing 2 |
| chr11_-_29658396 | 0.97 |
ENSDART00000183947
|
rpl22
|
ribosomal protein L22 |
| chr11_-_20071642 | 0.97 |
ENSDART00000162931
ENSDART00000159928 ENSDART00000191443 ENSDART00000121722 |
si:dkey-274m17.3
|
si:dkey-274m17.3 |
| chr5_-_52813442 | 0.96 |
ENSDART00000169305
|
zgc:158260
|
zgc:158260 |
| chr10_+_9159279 | 0.95 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr21_-_5393125 | 0.94 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr6_+_2097690 | 0.94 |
ENSDART00000193770
|
tgm2b
|
transglutaminase 2b |
| chr22_+_28969071 | 0.94 |
ENSDART00000163427
|
pimr95
|
Pim proto-oncogene, serine/threonine kinase, related 95 |
| chr3_+_11101585 | 0.94 |
ENSDART00000172103
|
sstr5
|
somatostatin receptor 5 |
| chr14_-_34513103 | 0.93 |
ENSDART00000136306
|
zgc:194246
|
zgc:194246 |
| chr15_-_45110011 | 0.93 |
ENSDART00000182047
ENSDART00000188662 |
CABZ01072607.1
|
|
| chr8_-_45760087 | 0.93 |
ENSDART00000025620
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr15_-_28095532 | 0.93 |
ENSDART00000191490
|
cryba1a
|
crystallin, beta A1a |
| chr6_-_2627488 | 0.93 |
ENSDART00000044089
ENSDART00000158333 ENSDART00000155109 |
hyi
|
hydroxypyruvate isomerase |
| chr13_+_24402406 | 0.93 |
ENSDART00000043002
|
rab1ab
|
RAB1A, member RAS oncogene family b |
| chr2_+_6126086 | 0.91 |
ENSDART00000179962
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr23_+_36771593 | 0.90 |
ENSDART00000078240
|
march9
|
membrane-associated ring finger (C3HC4) 9 |
| chr24_-_31306724 | 0.90 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr25_+_31868268 | 0.90 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr2_+_16160906 | 0.89 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr15_-_39948635 | 0.88 |
ENSDART00000114836
|
msh5
|
mutS homolog 5 |
| chr4_-_21652812 | 0.88 |
ENSDART00000174400
|
rps16
|
ribosomal protein S16 |
| chr7_+_65673885 | 0.88 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr18_+_12655766 | 0.88 |
ENSDART00000144246
|
tbxas1
|
thromboxane A synthase 1 (platelet) |
| chr13_+_37273010 | 0.88 |
ENSDART00000144387
|
gphb5
|
glycoprotein hormone beta 5 |
| chr2_+_19578446 | 0.87 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr12_+_695619 | 0.87 |
ENSDART00000161691
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr14_-_9056066 | 0.87 |
ENSDART00000139669
ENSDART00000138758 ENSDART00000041099 |
sybl1
|
synaptobrevin-like 1 |
| chr3_+_62196672 | 0.86 |
ENSDART00000097312
|
sco1
|
SCO1 cytochrome c oxidase assembly protein |
| chr12_+_47698356 | 0.86 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr4_-_779796 | 0.86 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr1_-_44899287 | 0.86 |
ENSDART00000187522
|
tcirg1a
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3a |
| chr7_-_35432901 | 0.85 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr4_+_27018663 | 0.85 |
ENSDART00000180778
|
CR749777.1
|
|
| chr1_-_45049603 | 0.84 |
ENSDART00000023336
|
rps6
|
ribosomal protein S6 |
| chr16_-_12512568 | 0.84 |
ENSDART00000055161
ENSDART00000160906 |
cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr4_+_22297839 | 0.82 |
ENSDART00000077707
|
llph
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr1_-_58562129 | 0.82 |
ENSDART00000159070
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr23_-_23401305 | 0.81 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr21_+_17956856 | 0.81 |
ENSDART00000080431
|
dnai1.2
|
dynein, axonemal, intermediate chain 1, paralog 2 |
| chr15_-_15469079 | 0.80 |
ENSDART00000132637
ENSDART00000004220 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr17_-_26926577 | 0.80 |
ENSDART00000050202
|
rcan3
|
regulator of calcineurin 3 |
| chr11_+_24815927 | 0.80 |
ENSDART00000146838
|
rabif
|
RAB interacting factor |
| chr1_-_36151377 | 0.79 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr13_+_51869025 | 0.79 |
ENSDART00000187066
|
LT631684.1
|
|
| chr10_-_28835771 | 0.79 |
ENSDART00000192220
ENSDART00000188436 |
alcama
|
activated leukocyte cell adhesion molecule a |
| chr16_+_53203370 | 0.79 |
ENSDART00000154669
|
si:ch211-269k10.2
|
si:ch211-269k10.2 |
| chr3_+_14388010 | 0.78 |
ENSDART00000171726
ENSDART00000165452 |
tmem56b
|
transmembrane protein 56b |
| chr15_+_11683114 | 0.78 |
ENSDART00000168233
|
si:dkey-31c13.1
|
si:dkey-31c13.1 |
| chr11_-_29657947 | 0.77 |
ENSDART00000125753
|
rpl22
|
ribosomal protein L22 |
| chr7_-_16596938 | 0.76 |
ENSDART00000134548
|
e2f8
|
E2F transcription factor 8 |
| chr11_-_41779281 | 0.76 |
ENSDART00000109204
|
megf6b
|
multiple EGF-like-domains 6b |
| chr11_+_24815667 | 0.76 |
ENSDART00000141730
|
rabif
|
RAB interacting factor |
| chr2_-_8611675 | 0.75 |
ENSDART00000138223
|
si:ch211-71m22.1
|
si:ch211-71m22.1 |
| chr19_+_935565 | 0.75 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr4_-_39111612 | 0.75 |
ENSDART00000150394
|
si:dkey-122c11.8
|
si:dkey-122c11.8 |
| chr23_+_4373360 | 0.74 |
ENSDART00000144061
|
ptpdc1b
|
protein tyrosine phosphatase domain containing 1b |
| chr14_+_30568961 | 0.74 |
ENSDART00000184303
|
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr2_-_45510223 | 0.73 |
ENSDART00000113058
|
gpsm2
|
G protein signaling modulator 2 |
| chr19_-_44089509 | 0.73 |
ENSDART00000189136
|
rad21b
|
RAD21 cohesin complex component b |
| chr4_+_2655358 | 0.73 |
ENSDART00000007638
|
bcap29
|
B cell receptor associated protein 29 |
| chr4_+_74141400 | 0.72 |
ENSDART00000166994
|
trim24
|
tripartite motif containing 24 |
| chr2_-_19520324 | 0.72 |
ENSDART00000079877
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr22_+_38978084 | 0.72 |
ENSDART00000025482
|
arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr12_+_16284086 | 0.72 |
ENSDART00000013360
ENSDART00000141169 |
ppp1r3cb
|
protein phosphatase 1, regulatory subunit 3Cb |
| chr18_-_6766354 | 0.71 |
ENSDART00000132611
|
adm2b
|
adrenomedullin 2b |
| chr17_+_35362851 | 0.71 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr11_+_37201483 | 0.71 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr20_+_53541389 | 0.71 |
ENSDART00000100110
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr8_-_48770163 | 0.70 |
ENSDART00000160959
|
pimr184
|
Pim proto-oncogene, serine/threonine kinase, related 184 |
| chr15_-_47956388 | 0.70 |
ENSDART00000116506
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr13_+_35635672 | 0.70 |
ENSDART00000148481
|
thbs2a
|
thrombospondin 2a |
| chr21_-_19314618 | 0.70 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr22_+_1796057 | 0.70 |
ENSDART00000170834
|
znf1179
|
zinc finger protein 1179 |
| chr25_+_34845115 | 0.69 |
ENSDART00000061996
|
tmem231
|
transmembrane protein 231 |
| chr23_+_32039386 | 0.69 |
ENSDART00000133801
|
mylk2
|
myosin light chain kinase 2 |
| chr1_-_524433 | 0.68 |
ENSDART00000147610
|
si:ch73-41e3.7
|
si:ch73-41e3.7 |
| chr11_-_45434959 | 0.67 |
ENSDART00000173106
ENSDART00000172767 ENSDART00000172933 ENSDART00000172986 |
rfc4
|
replication factor C (activator 1) 4 |
| chr7_+_21321317 | 0.67 |
ENSDART00000173950
|
dnah2
|
dynein, axonemal, heavy chain 2 |
| chr8_-_13064330 | 0.67 |
ENSDART00000129168
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr1_-_58561963 | 0.66 |
ENSDART00000165040
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr20_+_54274431 | 0.66 |
ENSDART00000099338
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr4_-_12914163 | 0.66 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr9_+_23748342 | 0.66 |
ENSDART00000019053
|
faima
|
Fas apoptotic inhibitory molecule a |
| chr12_-_47793857 | 0.65 |
ENSDART00000161294
|
dydc2
|
DPY30 domain containing 2 |
| chr22_-_7669624 | 0.65 |
ENSDART00000189617
|
AL929276.2
|
|
| chr13_+_9137116 | 0.65 |
ENSDART00000143293
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr4_+_25558849 | 0.65 |
ENSDART00000113663
ENSDART00000100755 ENSDART00000111416 ENSDART00000127840 ENSDART00000168618 ENSDART00000111820 ENSDART00000113866 ENSDART00000110107 ENSDART00000111344 ENSDART00000108548 |
zgc:195175
|
zgc:195175 |
| chr21_-_40676224 | 0.65 |
ENSDART00000162623
|
arxb
|
aristaless related homeobox b |
| chr1_+_16144615 | 0.64 |
ENSDART00000054707
|
tusc3
|
tumor suppressor candidate 3 |
| chr9_-_30555725 | 0.64 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr6_+_23931236 | 0.64 |
ENSDART00000166079
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr4_-_69189894 | 0.64 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr20_+_51199666 | 0.64 |
ENSDART00000169321
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr18_+_62932 | 0.63 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr25_+_36349574 | 0.63 |
ENSDART00000184101
|
zgc:173552
|
zgc:173552 |
| chr22_+_29067388 | 0.63 |
ENSDART00000133673
|
pimr100
|
Pim proto-oncogene, serine/threonine kinase, related 100 |
| chr12_+_9880493 | 0.62 |
ENSDART00000055019
|
ndufa4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 |
| chr7_+_61480296 | 0.62 |
ENSDART00000083255
|
adam19a
|
ADAM metallopeptidase domain 19a |
| chr25_+_16356083 | 0.62 |
ENSDART00000125925
ENSDART00000125444 |
tead1a
|
TEA domain family member 1a |
| chr18_-_14677936 | 0.62 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr8_-_21988833 | 0.62 |
ENSDART00000167708
|
nphp4
|
nephronophthisis 4 |
| chr9_-_54716363 | 0.62 |
ENSDART00000162158
ENSDART00000168430 |
trappc2
|
trafficking protein particle complex 2 |
| chr10_-_43771447 | 0.61 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr21_+_45502773 | 0.61 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr7_-_56831621 | 0.61 |
ENSDART00000182912
|
senp8
|
SUMO/sentrin peptidase family member, NEDD8 specific |
| chr3_-_62403550 | 0.61 |
ENSDART00000055055
|
sox8b
|
SRY (sex determining region Y)-box 8b |
| chr5_-_6508250 | 0.60 |
ENSDART00000060535
|
crybb3
|
crystallin, beta B3 |
| chr5_+_7279104 | 0.60 |
ENSDART00000190014
|
si:ch73-72b7.1
|
si:ch73-72b7.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of cux2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.4 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.7 | 2.9 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.5 | 1.6 | GO:0003097 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) |
| 0.5 | 1.4 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.4 | 1.6 | GO:0060306 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.4 | 1.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.4 | 1.1 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.4 | 1.8 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.3 | 1.0 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.3 | 1.0 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.3 | 1.5 | GO:0031650 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.3 | 0.8 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.3 | 1.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 1.7 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.2 | 0.7 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.2 | 0.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 0.6 | GO:0071634 | transforming growth factor beta activation(GO:0036363) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.2 | 2.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.6 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.2 | 0.6 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.2 | 1.1 | GO:0032728 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 0.5 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.2 | 0.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 0.8 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 2.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 1.7 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.2 | 0.9 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 2.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 2.0 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 2.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.5 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.9 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.7 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.5 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.6 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.1 | 0.4 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 1.5 | GO:0003314 | heart rudiment morphogenesis(GO:0003314) |
| 0.1 | 0.4 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.5 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 0.3 | GO:0036314 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.4 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.1 | 0.9 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 2.0 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.6 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.3 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 | 0.8 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 1.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.4 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.1 | 0.7 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 1.0 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 0.6 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.1 | 1.7 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 0.4 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 3.0 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.1 | 0.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 1.1 | GO:0051384 | response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) |
| 0.1 | 1.0 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 1.1 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.3 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.2 | GO:0044038 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 0.9 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.7 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.4 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 1.0 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 1.7 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.2 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.2 | GO:0043385 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.1 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.6 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.2 | GO:0060155 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.1 | 0.4 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.1 | 0.6 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.3 | GO:2000379 | protein import into mitochondrial inner membrane(GO:0045039) positive regulation of reactive oxygen species metabolic process(GO:2000379) |
| 0.1 | 0.7 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.7 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 0.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.3 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 0.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.9 | GO:0048771 | tissue remodeling(GO:0048771) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.9 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.2 | GO:0016122 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 0.5 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.4 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.3 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.1 | 0.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.4 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.2 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.9 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 1.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 2.2 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.3 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.3 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.2 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 1.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0014896 | muscle hypertrophy(GO:0014896) |
| 0.0 | 0.6 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.2 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.7 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.4 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.1 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.1 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.0 | 0.2 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:0046635 | positive regulation of myeloid leukocyte mediated immunity(GO:0002888) positive regulation of alpha-beta T cell activation(GO:0046635) |
| 0.0 | 0.3 | GO:0021781 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) glial cell fate commitment(GO:0021781) |
| 0.0 | 0.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.3 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.5 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 2.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.7 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.1 | GO:0046144 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.0 | 0.3 | GO:1902547 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.0 | 0.4 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.3 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.5 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.0 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.9 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 1.5 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 1.3 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 2.3 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.0 | 0.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.0 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:1902259 | regulation of potassium ion transmembrane transporter activity(GO:1901016) regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.9 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.8 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.1 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.3 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.2 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.4 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.8 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 1.0 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.2 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 1.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.5 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.2 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.3 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 1.4 | GO:0032880 | regulation of protein localization(GO:0032880) |
| 0.0 | 0.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.2 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 4.5 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.0 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.0 | 0.7 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 1.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.3 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.9 | GO:0010970 | establishment of localization by movement along microtubule(GO:0010970) |
| 0.0 | 0.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0048641 | regulation of skeletal muscle tissue development(GO:0048641) |
| 0.0 | 0.0 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 1.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) commissural neuron axon guidance(GO:0071679) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.3 | 1.6 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 2.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 2.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 1.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 0.6 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 1.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 1.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.8 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 1.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.7 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.4 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.6 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 0.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.2 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 0.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 0.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 3.3 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 2.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.3 | GO:0032426 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 2.0 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 3.8 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.4 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 2.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.5 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.3 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.0 | 0.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.4 | GO:0090544 | BAF-type complex(GO:0090544) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0031834 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.4 | 1.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.4 | 2.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.4 | 2.9 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.3 | 1.0 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.3 | 1.3 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.3 | 3.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.2 | 0.7 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 1.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 1.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.9 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 1.6 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.2 | 1.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 0.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.2 | 2.5 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 0.5 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.2 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 1.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 1.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 3.9 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.7 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 1.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.8 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.7 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 1.9 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.9 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.9 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 1.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 2.0 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.5 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.8 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 1.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 3.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.6 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.3 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.1 | 0.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.3 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 1.5 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 0.2 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.1 | 0.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.2 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 0.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 1.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.3 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.9 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 0.3 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 1.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.9 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.5 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.3 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 1.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 7.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.0 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 1.1 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 3.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 2.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 5.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 1.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 1.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.6 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 1.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.5 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 2.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.4 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 1.9 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 2.0 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 3.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 0.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 1.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 3.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.5 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |