Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for cux1a
Z-value: 0.09
Transcription factors associated with cux1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cux1a
|
ENSDARG00000078459 | cut-like homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cux1a | dr11_v1_chr10_-_33156789_33156789 | -0.54 | 2.2e-02 | Click! |
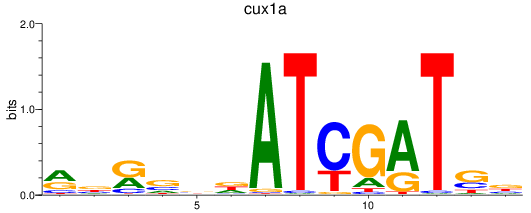
Activity profile of cux1a motif
Sorted Z-values of cux1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_7607114 | 0.25 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr18_+_7611298 | 0.23 |
ENSDART00000062156
|
odf3b
|
outer dense fiber of sperm tails 3B |
| chr22_-_15562933 | 0.21 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr16_+_25126935 | 0.18 |
ENSDART00000058945
|
zgc:92590
|
zgc:92590 |
| chr17_+_12159529 | 0.16 |
ENSDART00000046802
|
sccpdha
|
saccharopine dehydrogenase a |
| chr23_-_33986762 | 0.15 |
ENSDART00000144609
|
tmed4
|
transmembrane p24 trafficking protein 4 |
| chr12_+_48220584 | 0.15 |
ENSDART00000164392
|
lrrc20
|
leucine rich repeat containing 20 |
| chr22_+_29067388 | 0.12 |
ENSDART00000133673
|
pimr100
|
Pim proto-oncogene, serine/threonine kinase, related 100 |
| chr25_-_6082509 | 0.11 |
ENSDART00000104755
|
cpeb1a
|
cytoplasmic polyadenylation element binding protein 1a |
| chr15_+_36966369 | 0.11 |
ENSDART00000163622
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr5_-_13835461 | 0.11 |
ENSDART00000148297
ENSDART00000114841 |
add2
|
adducin 2 (beta) |
| chr22_+_28969071 | 0.10 |
ENSDART00000163427
|
pimr95
|
Pim proto-oncogene, serine/threonine kinase, related 95 |
| chr2_+_7715810 | 0.10 |
ENSDART00000130781
|
eif4a2
|
eukaryotic translation initiation factor 4A, isoform 2 |
| chr20_+_33875256 | 0.09 |
ENSDART00000002554
|
rxrgb
|
retinoid X receptor, gamma b |
| chr16_+_19014886 | 0.09 |
ENSDART00000079298
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr25_-_35182347 | 0.09 |
ENSDART00000115210
|
ano9a
|
anoctamin 9a |
| chr11_-_12673637 | 0.08 |
ENSDART00000186058
|
CR450764.7
|
|
| chr21_+_21012210 | 0.08 |
ENSDART00000190317
|
ndr1
|
nodal-related 1 |
| chr18_+_5490668 | 0.08 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr22_+_13917311 | 0.08 |
ENSDART00000022654
|
sh3bp4a
|
SH3-domain binding protein 4a |
| chrM_+_9735 | 0.07 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr11_-_12705608 | 0.07 |
ENSDART00000158937
ENSDART00000114391 |
zgc:174353
|
zgc:174353 |
| chr12_-_19091214 | 0.07 |
ENSDART00000153225
|
si:ch73-139e5.4
|
si:ch73-139e5.4 |
| chr20_-_54075136 | 0.07 |
ENSDART00000074255
|
mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr11_+_16152316 | 0.07 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr7_+_49272702 | 0.06 |
ENSDART00000083389
|
abtb2a
|
ankyrin repeat and BTB (POZ) domain containing 2a |
| chr8_+_48491387 | 0.06 |
ENSDART00000086053
|
PRDM16
|
si:ch211-263k4.2 |
| chr4_-_14915268 | 0.06 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr7_-_25895189 | 0.06 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr16_+_1383914 | 0.06 |
ENSDART00000185089
|
cers2b
|
ceramide synthase 2b |
| chr7_+_29951997 | 0.06 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr4_-_25485404 | 0.05 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr24_+_35881124 | 0.05 |
ENSDART00000143015
|
klhl14
|
kelch-like family member 14 |
| chr25_+_21324588 | 0.05 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr3_-_37082618 | 0.05 |
ENSDART00000026701
ENSDART00000110716 |
tubg1
|
tubulin, gamma 1 |
| chr22_+_34691020 | 0.04 |
ENSDART00000192202
|
hykk.1
|
hydroxylysine kinase, tandem duplicate 1 |
| chr10_-_11397590 | 0.04 |
ENSDART00000064212
|
srek1ip1
|
SREK1-interacting protein 1 |
| chr6_-_29105727 | 0.04 |
ENSDART00000184355
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr23_+_3721042 | 0.04 |
ENSDART00000143323
|
smim29
|
small integral membrane protein 29 |
| chr24_-_39826865 | 0.04 |
ENSDART00000089232
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr11_-_16152400 | 0.04 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr23_-_44494401 | 0.03 |
ENSDART00000114640
ENSDART00000148532 |
actl6b
|
actin-like 6B |
| chr15_+_42626957 | 0.03 |
ENSDART00000154754
|
grik1b
|
glutamate receptor, ionotropic, kainate 1b |
| chr23_+_9353552 | 0.03 |
ENSDART00000163298
|
BX511246.1
|
|
| chr12_-_9700605 | 0.03 |
ENSDART00000161063
|
heatr1
|
HEAT repeat containing 1 |
| chr16_+_4658250 | 0.03 |
ENSDART00000006212
|
LIN28A
|
si:ch1073-284b18.2 |
| chr3_-_5555500 | 0.03 |
ENSDART00000176133
ENSDART00000057464 |
trim35-31
|
tripartite motif containing 35-31 |
| chr11_-_16152105 | 0.03 |
ENSDART00000081062
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr17_+_53435279 | 0.03 |
ENSDART00000126630
|
cx52.9
|
connexin 52.9 |
| chr21_+_17768174 | 0.02 |
ENSDART00000141380
|
rxraa
|
retinoid X receptor, alpha a |
| chr1_-_49932040 | 0.02 |
ENSDART00000048281
|
cyp2u1
|
cytochrome P450, family 2, subfamily U, polypeptide 1 |
| chr8_-_29851706 | 0.02 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr23_-_24047054 | 0.02 |
ENSDART00000184308
ENSDART00000185902 |
CR925720.1
|
|
| chr1_+_55703120 | 0.02 |
ENSDART00000141089
|
adgre6
|
adhesion G protein-coupled receptor E6 |
| chr14_-_25985698 | 0.02 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr22_+_28446365 | 0.02 |
ENSDART00000189359
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr4_-_51460013 | 0.02 |
ENSDART00000193382
|
CR628395.1
|
|
| chr17_+_45454943 | 0.02 |
ENSDART00000074838
|
kcnk3b
|
potassium channel, subfamily K, member 3b |
| chr12_-_4185554 | 0.02 |
ENSDART00000152313
|
si:dkey-32n7.7
|
si:dkey-32n7.7 |
| chr24_-_3407507 | 0.01 |
ENSDART00000132648
|
nck1b
|
NCK adaptor protein 1b |
| chr17_+_11507131 | 0.01 |
ENSDART00000013170
|
kif26ba
|
kinesin family member 26Ba |
| chr17_-_30839338 | 0.01 |
ENSDART00000139707
|
gdf7
|
growth differentiation factor 7 |
| chr17_+_48164536 | 0.01 |
ENSDART00000161750
ENSDART00000156923 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr3_-_38692920 | 0.01 |
ENSDART00000155042
|
mpp3a
|
membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) |
| chr20_+_32406011 | 0.01 |
ENSDART00000018640
ENSDART00000137910 |
snx3
|
sorting nexin 3 |
| chr3_-_31079186 | 0.01 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr22_+_28446557 | 0.01 |
ENSDART00000089546
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr16_-_24561354 | 0.01 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr2_-_56649883 | 0.01 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr8_-_24135171 | 0.01 |
ENSDART00000112926
|
adora1b
|
adenosine A1 receptor b |
| chr12_-_4540564 | 0.00 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr16_-_17175731 | 0.00 |
ENSDART00000183057
|
opn9
|
opsin 9 |
| chr1_-_39976492 | 0.00 |
ENSDART00000181680
|
stox2a
|
storkhead box 2a |
| chr15_-_16704417 | 0.00 |
ENSDART00000155163
|
caln1
|
calneuron 1 |
| chr13_-_36264861 | 0.00 |
ENSDART00000100204
|
slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr10_+_21786656 | 0.00 |
ENSDART00000185851
ENSDART00000167219 |
pcdh1g26
|
protocadherin 1 gamma 26 |
| chr11_-_20956309 | 0.00 |
ENSDART00000188659
|
CABZ01008739.1
|
|
| chr16_-_12095144 | 0.00 |
ENSDART00000145106
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr19_+_14059349 | 0.00 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr14_-_33083539 | 0.00 |
ENSDART00000160173
|
dlg3
|
discs, large homolog 3 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of cux1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |