Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
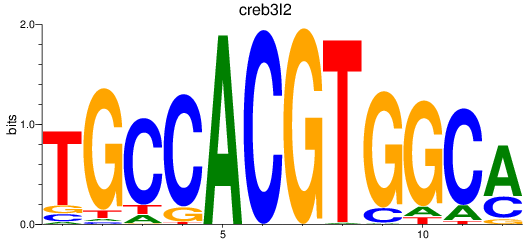
Results for creb3l2
Z-value: 0.38
Transcription factors associated with creb3l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
creb3l2
|
ENSDARG00000063563 | cAMP responsive element binding protein 3-like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| creb3l2 | dr11_v1_chr4_-_4751981_4751983 | 0.55 | 1.8e-02 | Click! |
Activity profile of creb3l2 motif
Sorted Z-values of creb3l2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_17200120 | 0.82 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr22_-_817479 | 0.57 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr7_+_38716048 | 0.57 |
ENSDART00000024590
|
syt13
|
synaptotagmin XIII |
| chr5_-_22130937 | 0.52 |
ENSDART00000138606
|
las1l
|
LAS1-like, ribosome biogenesis factor |
| chr5_-_54714525 | 0.48 |
ENSDART00000150138
ENSDART00000150070 |
ccnb1
|
cyclin B1 |
| chr25_+_34862225 | 0.44 |
ENSDART00000149782
|
CHST6
|
zgc:194879 |
| chr5_-_54714789 | 0.44 |
ENSDART00000063357
|
ccnb1
|
cyclin B1 |
| chr18_-_43857089 | 0.44 |
ENSDART00000150170
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr19_+_7173613 | 0.43 |
ENSDART00000001331
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr16_+_22865942 | 0.41 |
ENSDART00000103235
ENSDART00000143957 |
flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr5_-_32338866 | 0.40 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr18_-_37252036 | 0.40 |
ENSDART00000132230
|
six5
|
SIX homeobox 5 |
| chr14_+_22076596 | 0.39 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr15_-_43978141 | 0.39 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr20_-_13140309 | 0.37 |
ENSDART00000020703
ENSDART00000188594 |
ints7
|
integrator complex subunit 7 |
| chr15_+_32419303 | 0.37 |
ENSDART00000162663
|
si:dkey-285b23.3
|
si:dkey-285b23.3 |
| chr5_+_69278089 | 0.36 |
ENSDART00000136656
|
selenom
|
selenoprotein M |
| chr1_+_2301961 | 0.35 |
ENSDART00000108919
ENSDART00000143361 ENSDART00000142944 |
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr7_-_69352424 | 0.34 |
ENSDART00000170714
|
ap1g1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr20_+_51061695 | 0.33 |
ENSDART00000134416
|
im:7140055
|
im:7140055 |
| chr16_+_20294976 | 0.33 |
ENSDART00000059619
|
fkbp14
|
FK506 binding protein 14 |
| chr13_+_36958086 | 0.32 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr5_-_63644938 | 0.32 |
ENSDART00000050865
|
surf4l
|
surfeit gene 4, like |
| chr5_-_67757188 | 0.31 |
ENSDART00000167168
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr16_-_32727199 | 0.31 |
ENSDART00000137232
|
usp45
|
ubiquitin specific peptidase 45 |
| chr9_+_31222026 | 0.30 |
ENSDART00000145573
|
clybl
|
citrate lyase beta like |
| chr25_-_1323623 | 0.29 |
ENSDART00000156532
ENSDART00000157163 ENSDART00000156062 ENSDART00000082447 ENSDART00000189175 |
calml4b
|
calmodulin-like 4b |
| chr25_+_3788443 | 0.28 |
ENSDART00000189747
|
chid1
|
chitinase domain containing 1 |
| chr13_-_25548733 | 0.28 |
ENSDART00000168099
ENSDART00000135788 ENSDART00000077655 |
mcmbp
|
minichromosome maintenance complex binding protein |
| chr4_+_77957611 | 0.27 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr25_-_7670616 | 0.26 |
ENSDART00000131583
ENSDART00000142794 |
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr16_+_813780 | 0.26 |
ENSDART00000162474
ENSDART00000161774 |
irx1a
|
iroquois homeobox 1a |
| chr17_+_44441042 | 0.26 |
ENSDART00000142123
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr8_-_46386024 | 0.21 |
ENSDART00000136602
ENSDART00000060919 ENSDART00000137472 |
qars
|
glutaminyl-tRNA synthetase |
| chr4_+_17844013 | 0.21 |
ENSDART00000019165
|
apaf1
|
apoptotic peptidase activating factor 1 |
| chr12_+_4220353 | 0.21 |
ENSDART00000133675
|
mapk7
|
mitogen-activated protein kinase 7 |
| chr8_-_46045172 | 0.20 |
ENSDART00000135138
|
si:ch211-119d14.3
|
si:ch211-119d14.3 |
| chr13_+_1015749 | 0.20 |
ENSDART00000190982
|
prokr1b
|
prokineticin receptor 1b |
| chr8_+_7316568 | 0.20 |
ENSDART00000140874
|
selenoh
|
selenoprotein H |
| chr6_-_23931442 | 0.20 |
ENSDART00000160547
|
sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr17_-_44440832 | 0.18 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr2_-_11504347 | 0.17 |
ENSDART00000019392
|
sdr16c5a
|
short chain dehydrogenase/reductase family 16C, member 5a |
| chr3_+_42923275 | 0.17 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr2_-_10386738 | 0.17 |
ENSDART00000016369
|
wls
|
wntless Wnt ligand secretion mediator |
| chr4_+_17843717 | 0.17 |
ENSDART00000113507
|
apaf1
|
apoptotic peptidase activating factor 1 |
| chr14_+_16287968 | 0.17 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr8_+_49065348 | 0.17 |
ENSDART00000032277
|
ddx51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr17_-_28797395 | 0.16 |
ENSDART00000134735
|
scfd1
|
sec1 family domain containing 1 |
| chr3_+_53773256 | 0.15 |
ENSDART00000170461
|
col5a3a
|
collagen, type V, alpha 3a |
| chr12_+_19408373 | 0.15 |
ENSDART00000114248
|
snx29
|
sorting nexin 29 |
| chr3_-_18384501 | 0.15 |
ENSDART00000027630
|
kdelr2a
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2a |
| chr2_+_50999675 | 0.15 |
ENSDART00000158064
ENSDART00000165746 ENSDART00000163917 ENSDART00000172038 ENSDART00000169048 ENSDART00000164775 |
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr18_-_13056801 | 0.12 |
ENSDART00000088908
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr2_+_50999477 | 0.12 |
ENSDART00000190111
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr20_+_29209767 | 0.12 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr22_-_12337781 | 0.12 |
ENSDART00000188357
ENSDART00000123574 |
zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr6_+_42693288 | 0.12 |
ENSDART00000155010
|
si:ch211-207d10.2
|
si:ch211-207d10.2 |
| chr3_+_39568290 | 0.11 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr6_+_42693114 | 0.11 |
ENSDART00000154353
|
si:ch211-207d10.2
|
si:ch211-207d10.2 |
| chr2_-_39015130 | 0.11 |
ENSDART00000044331
|
copb2
|
coatomer protein complex, subunit beta 2 |
| chr2_-_44971551 | 0.11 |
ENSDART00000018818
|
mul1a
|
mitochondrial E3 ubiquitin protein ligase 1a |
| chr20_+_29209926 | 0.10 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr21_+_44857293 | 0.10 |
ENSDART00000134365
ENSDART00000168217 ENSDART00000065083 |
fstl4
|
follistatin-like 4 |
| chr22_+_31059919 | 0.10 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr22_-_25043103 | 0.10 |
ENSDART00000015512
|
polr3f
|
polymerase (RNA) III (DNA directed) polypeptide F |
| chr4_-_22749553 | 0.09 |
ENSDART00000040033
|
nup107
|
nucleoporin 107 |
| chr5_-_28968964 | 0.09 |
ENSDART00000184936
ENSDART00000016628 |
fam129bb
|
family with sequence similarity 129, member Bb |
| chr1_-_26444075 | 0.08 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr22_+_25841786 | 0.08 |
ENSDART00000180863
|
vasna
|
vasorin a |
| chr11_+_25296366 | 0.08 |
ENSDART00000065949
|
top1l
|
DNA topoisomerase I, like |
| chr25_-_37084032 | 0.08 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr20_+_15565926 | 0.07 |
ENSDART00000063917
|
si:dkey-86e18.1
|
si:dkey-86e18.1 |
| chr8_+_8699085 | 0.07 |
ENSDART00000021209
|
uxt
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr21_-_5881344 | 0.07 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr21_+_21357309 | 0.07 |
ENSDART00000079634
ENSDART00000146102 |
kcnk12l
|
potassium channel, subfamily K, member 12 like |
| chr11_-_7156620 | 0.07 |
ENSDART00000172823
ENSDART00000172879 ENSDART00000078916 |
smim7
|
small integral membrane protein 7 |
| chr7_+_15308042 | 0.06 |
ENSDART00000185170
|
mespaa
|
mesoderm posterior aa |
| chr14_-_3071564 | 0.06 |
ENSDART00000166136
ENSDART00000167138 |
slc35a4
|
solute carrier family 35, member A4 |
| chr6_+_269204 | 0.05 |
ENSDART00000191678
|
atf4a
|
activating transcription factor 4a |
| chr15_+_32249062 | 0.05 |
ENSDART00000133867
ENSDART00000152545 ENSDART00000082315 ENSDART00000152513 ENSDART00000152139 |
arfip2a
|
ADP-ribosylation factor interacting protein 2a |
| chr15_-_12229874 | 0.05 |
ENSDART00000165159
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr24_+_32472155 | 0.04 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr11_+_45323455 | 0.04 |
ENSDART00000189249
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr22_+_9871238 | 0.04 |
ENSDART00000141085
ENSDART00000105939 |
si:dkey-253d23.4
|
si:dkey-253d23.4 |
| chr21_-_3613702 | 0.04 |
ENSDART00000139194
|
dym
|
dymeclin |
| chr10_-_7913591 | 0.04 |
ENSDART00000139661
|
slc35e4
|
solute carrier family 35, member E4 |
| chr4_-_77227748 | 0.03 |
ENSDART00000171545
|
si:dkey-172k15.4
|
si:dkey-172k15.4 |
| chr10_-_1180314 | 0.03 |
ENSDART00000114261
|
bmpr1bb
|
bone morphogenetic protein receptor, type IBb |
| chr16_+_33109496 | 0.03 |
ENSDART00000162364
|
cnr2
|
cannabinoid receptor 2 |
| chr14_-_32016615 | 0.03 |
ENSDART00000105761
|
zic3
|
zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
| chr7_-_47148440 | 0.03 |
ENSDART00000185435
ENSDART00000189719 |
BX005073.3
|
|
| chr4_-_17391091 | 0.02 |
ENSDART00000056002
|
th2
|
tyrosine hydroxylase 2 |
| chr2_+_39618951 | 0.02 |
ENSDART00000077108
|
zgc:136870
|
zgc:136870 |
| chr14_+_46028003 | 0.02 |
ENSDART00000113469
|
nocta
|
nocturnin a |
| chr24_+_12913329 | 0.01 |
ENSDART00000141829
|
si:dkeyp-28d2.4
|
si:dkeyp-28d2.4 |
| chr15_+_897280 | 0.01 |
ENSDART00000155470
|
znf1012
|
zinc finger protein 1012 |
| chr4_-_12718103 | 0.01 |
ENSDART00000144388
|
mgst1.1
|
microsomal glutathione S-transferase 1.1 |
| chr21_+_45685757 | 0.01 |
ENSDART00000160530
|
sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr22_-_94352 | 0.01 |
ENSDART00000184883
|
ndufaf3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr7_-_45990386 | 0.01 |
ENSDART00000186008
|
si:ch211-260e23.7
|
si:ch211-260e23.7 |
| chr9_+_42095220 | 0.01 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr13_-_4992395 | 0.00 |
ENSDART00000102651
|
nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr1_+_17593392 | 0.00 |
ENSDART00000078889
|
helt
|
helt bHLH transcription factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of creb3l2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.1 | 0.4 | GO:0006747 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.1 | 0.9 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.3 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.2 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 0.4 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.2 | GO:0070973 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.8 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.3 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.2 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.3 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.6 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.3 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.4 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.1 | 0.4 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.1 | 0.3 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.1 | 0.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.8 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |