Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
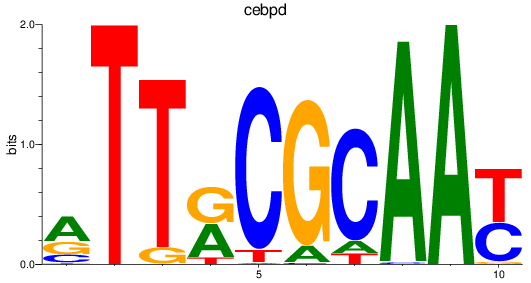
Results for cebpd
Z-value: 0.59
Transcription factors associated with cebpd
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebpd
|
ENSDARG00000087303 | CCAAT enhancer binding protein delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cebpd | dr11_v1_chr24_+_35564668_35564668 | -0.30 | 2.3e-01 | Click! |
Activity profile of cebpd motif
Sorted Z-values of cebpd motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_46020508 | 2.06 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr7_+_46019780 | 1.79 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr5_+_22974019 | 1.69 |
ENSDART00000147157
ENSDART00000020434 |
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr6_+_10333920 | 1.45 |
ENSDART00000151667
ENSDART00000151477 |
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr19_-_25113660 | 1.41 |
ENSDART00000035538
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr12_+_4920451 | 1.27 |
ENSDART00000171525
ENSDART00000159986 |
plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr6_+_23810529 | 1.19 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr5_-_15283509 | 1.16 |
ENSDART00000052712
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr13_+_40437550 | 1.13 |
ENSDART00000057090
ENSDART00000167859 |
got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr21_+_17051478 | 1.09 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr6_+_44197348 | 1.06 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr6_+_44197099 | 1.03 |
ENSDART00000124168
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr6_+_11397269 | 1.00 |
ENSDART00000114260
|
senp2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr7_+_39418869 | 0.89 |
ENSDART00000169195
|
CT030188.1
|
|
| chr19_-_20446756 | 0.89 |
ENSDART00000140711
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr9_-_53062083 | 0.88 |
ENSDART00000122155
|
zmp:0000000936
|
zmp:0000000936 |
| chr7_+_67733759 | 0.84 |
ENSDART00000172015
|
cyb5b
|
cytochrome b5 type B |
| chr14_-_5642371 | 0.83 |
ENSDART00000183859
ENSDART00000054876 |
npm1b
|
nucleophosmin 1b |
| chr8_+_42917515 | 0.83 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr6_+_8314451 | 0.81 |
ENSDART00000147793
ENSDART00000183688 |
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr19_+_43341115 | 0.81 |
ENSDART00000145846
ENSDART00000102384 |
sesn2
|
sestrin 2 |
| chr18_-_15551360 | 0.80 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr14_+_21699129 | 0.78 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr19_+_43341424 | 0.78 |
ENSDART00000134815
|
sesn2
|
sestrin 2 |
| chr11_+_13176568 | 0.77 |
ENSDART00000125371
ENSDART00000123257 |
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr9_+_24065855 | 0.76 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr6_+_8315050 | 0.73 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr5_+_8196264 | 0.73 |
ENSDART00000174564
ENSDART00000161261 |
lmbrd2a
|
LMBR1 domain containing 2a |
| chr9_-_9225980 | 0.72 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr5_-_54395488 | 0.72 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr18_+_7553950 | 0.72 |
ENSDART00000193420
ENSDART00000062143 |
zgc:77650
|
zgc:77650 |
| chr5_+_51111343 | 0.72 |
ENSDART00000092002
|
pomt1
|
protein-O-mannosyltransferase 1 |
| chr16_-_27138478 | 0.71 |
ENSDART00000147438
|
tmem245
|
transmembrane protein 245 |
| chr16_-_7793457 | 0.70 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr18_-_37252036 | 0.69 |
ENSDART00000132230
|
six5
|
SIX homeobox 5 |
| chr13_+_30035253 | 0.69 |
ENSDART00000181303
ENSDART00000057525 ENSDART00000136622 |
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr5_+_51111766 | 0.69 |
ENSDART00000188552
|
pomt1
|
protein-O-mannosyltransferase 1 |
| chr8_+_26874924 | 0.68 |
ENSDART00000141794
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr17_-_17759138 | 0.66 |
ENSDART00000157128
ENSDART00000123845 |
adck1
|
aarF domain containing kinase 1 |
| chr5_-_69948099 | 0.65 |
ENSDART00000034639
ENSDART00000191111 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr19_-_15192638 | 0.63 |
ENSDART00000048151
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr14_+_21699414 | 0.63 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr12_+_13205955 | 0.63 |
ENSDART00000092906
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr20_-_33675676 | 0.61 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr8_-_11324143 | 0.61 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr2_-_54387550 | 0.61 |
ENSDART00000097388
|
napgb
|
N-ethylmaleimide-sensitive factor attachment protein, gamma b |
| chr14_+_3507326 | 0.60 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr2_-_4032732 | 0.60 |
ENSDART00000158335
|
rab18b
|
RAB18B, member RAS oncogene family |
| chr4_-_4261673 | 0.60 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr2_-_37277626 | 0.59 |
ENSDART00000135340
|
nadkb
|
NAD kinase b |
| chr12_-_17592215 | 0.59 |
ENSDART00000134597
|
usp42
|
ubiquitin specific peptidase 42 |
| chr16_-_9830451 | 0.58 |
ENSDART00000148528
|
grhl2a
|
grainyhead-like transcription factor 2a |
| chr5_-_48680580 | 0.57 |
ENSDART00000031194
|
lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr5_-_23574234 | 0.57 |
ENSDART00000002453
|
cwc15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr9_-_31915423 | 0.56 |
ENSDART00000060051
|
fgf14
|
fibroblast growth factor 14 |
| chr5_-_43682930 | 0.56 |
ENSDART00000075017
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr16_-_42770064 | 0.54 |
ENSDART00000112879
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr10_+_45248326 | 0.54 |
ENSDART00000159797
|
zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr2_-_7246848 | 0.53 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr24_-_31123365 | 0.53 |
ENSDART00000182947
|
tmem56a
|
transmembrane protein 56a |
| chr25_-_12805295 | 0.52 |
ENSDART00000157629
|
ca5a
|
carbonic anhydrase Va |
| chr22_-_16377960 | 0.51 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr22_-_16377666 | 0.49 |
ENSDART00000161878
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr22_-_10158038 | 0.47 |
ENSDART00000047444
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr11_+_43751263 | 0.47 |
ENSDART00000163843
|
zgc:153431
|
zgc:153431 |
| chr15_+_24676905 | 0.46 |
ENSDART00000078014
ENSDART00000143137 |
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr19_-_37508571 | 0.46 |
ENSDART00000018255
|
ilf2
|
interleukin enhancer binding factor 2 |
| chr23_+_38957472 | 0.46 |
ENSDART00000193836
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr11_+_31323746 | 0.46 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr8_+_17884569 | 0.45 |
ENSDART00000134660
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr8_-_35960987 | 0.45 |
ENSDART00000160503
|
slc15a4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr7_-_37895771 | 0.44 |
ENSDART00000084282
|
papd5
|
PAP associated domain containing 5 |
| chr20_-_35508805 | 0.41 |
ENSDART00000169538
|
adgrf3b
|
adhesion G protein-coupled receptor F3b |
| chr21_+_30721733 | 0.41 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr16_-_7828838 | 0.41 |
ENSDART00000191434
ENSDART00000108653 |
tcaim
|
T cell activation inhibitor, mitochondrial |
| chr14_-_12071679 | 0.40 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr12_-_22238004 | 0.40 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr13_-_30996072 | 0.39 |
ENSDART00000181661
|
wdfy4
|
WDFY family member 4 |
| chr17_+_43013171 | 0.39 |
ENSDART00000055541
|
gskip
|
gsk3b interacting protein |
| chr12_+_17154655 | 0.36 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr6_-_10912424 | 0.35 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr11_+_23933016 | 0.35 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr13_-_32898962 | 0.35 |
ENSDART00000163757
|
rock2a
|
rho-associated, coiled-coil containing protein kinase 2a |
| chr3_+_15394428 | 0.34 |
ENSDART00000133168
|
atxn2l
|
ataxin 2-like |
| chr6_-_10728057 | 0.34 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr15_-_29348212 | 0.33 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr9_+_2574122 | 0.33 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr14_+_16345003 | 0.31 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr13_-_25548733 | 0.30 |
ENSDART00000168099
ENSDART00000135788 ENSDART00000077655 |
mcmbp
|
minichromosome maintenance complex binding protein |
| chr7_+_38089650 | 0.29 |
ENSDART00000052365
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr12_+_34953038 | 0.29 |
ENSDART00000187022
ENSDART00000123988 ENSDART00000027034 |
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr3_+_41731527 | 0.27 |
ENSDART00000049007
ENSDART00000187866 |
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr12_+_36109507 | 0.27 |
ENSDART00000175409
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr22_+_15973122 | 0.27 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr9_-_32300783 | 0.27 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr6_-_14292307 | 0.26 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr12_-_26383242 | 0.26 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr23_-_36003441 | 0.25 |
ENSDART00000164699
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr17_-_10838434 | 0.25 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr3_+_36972586 | 0.24 |
ENSDART00000102784
|
si:ch211-18i17.2
|
si:ch211-18i17.2 |
| chr3_+_19216567 | 0.24 |
ENSDART00000134433
|
il12rb2l
|
interleukin 12 receptor, beta 2a, like |
| chr23_-_36003282 | 0.23 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr19_-_30810328 | 0.22 |
ENSDART00000184875
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr5_-_26893310 | 0.22 |
ENSDART00000126669
|
lman2lb
|
lectin, mannose-binding 2-like b |
| chr14_+_7902374 | 0.22 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr12_+_32368574 | 0.21 |
ENSDART00000086389
|
ANKFN1
|
si:ch211-277e21.2 |
| chr10_+_44940693 | 0.21 |
ENSDART00000157515
|
cnnm4a
|
cyclin and CBS domain divalent metal cation transport mediator 4a |
| chr20_-_19511700 | 0.20 |
ENSDART00000040191
|
snx17
|
sorting nexin 17 |
| chr21_+_29227224 | 0.19 |
ENSDART00000013641
|
adra1ba
|
adrenoceptor alpha 1Ba |
| chr17_+_23975762 | 0.19 |
ENSDART00000155941
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr24_+_9881219 | 0.18 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr3_+_39663987 | 0.18 |
ENSDART00000184614
ENSDART00000184573 ENSDART00000183127 |
epn2
|
epsin 2 |
| chr3_-_30153242 | 0.17 |
ENSDART00000077089
|
nucb1
|
nucleobindin 1 |
| chr9_-_32300611 | 0.16 |
ENSDART00000127938
|
hspd1
|
heat shock 60 protein 1 |
| chr15_-_24960730 | 0.16 |
ENSDART00000109990
ENSDART00000186706 |
abhd15a
|
abhydrolase domain containing 15a |
| chr8_-_37043900 | 0.16 |
ENSDART00000139567
|
renbp
|
renin binding protein |
| chr2_+_11029138 | 0.15 |
ENSDART00000138737
ENSDART00000081058 ENSDART00000153662 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr7_-_22790630 | 0.15 |
ENSDART00000173496
|
si:ch211-15b10.6
|
si:ch211-15b10.6 |
| chr24_-_32665283 | 0.14 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr3_-_30152836 | 0.14 |
ENSDART00000165920
|
nucb1
|
nucleobindin 1 |
| chr21_-_14762944 | 0.14 |
ENSDART00000114096
|
arrdc1b
|
arrestin domain containing 1b |
| chr5_+_36439405 | 0.14 |
ENSDART00000102973
|
eda
|
ectodysplasin A |
| chr25_-_16987020 | 0.14 |
ENSDART00000073419
|
ccnd2a
|
cyclin D2, a |
| chr4_+_33012407 | 0.14 |
ENSDART00000151873
|
si:dkey-26h11.2
|
si:dkey-26h11.2 |
| chr9_-_12034444 | 0.13 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr17_+_5623514 | 0.12 |
ENSDART00000171220
ENSDART00000176083 |
CU571310.1
|
|
| chr15_+_21882419 | 0.12 |
ENSDART00000157216
|
si:dkey-103g5.4
|
si:dkey-103g5.4 |
| chr11_-_29082175 | 0.12 |
ENSDART00000123245
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr3_+_36972298 | 0.11 |
ENSDART00000150917
|
si:ch211-18i17.2
|
si:ch211-18i17.2 |
| chr7_-_39751540 | 0.11 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr25_+_3294150 | 0.10 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr8_-_36412936 | 0.09 |
ENSDART00000159276
|
si:zfos-2070c2.3
|
si:zfos-2070c2.3 |
| chr7_-_7904683 | 0.09 |
ENSDART00000192822
|
CABZ01030278.1
|
|
| chr10_+_9595575 | 0.09 |
ENSDART00000091780
ENSDART00000184287 |
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr2_-_49860723 | 0.09 |
ENSDART00000083690
|
blvra
|
biliverdin reductase A |
| chr3_-_43821191 | 0.09 |
ENSDART00000186816
|
snn
|
stannin |
| chr22_+_31244728 | 0.08 |
ENSDART00000163309
|
grip2b
|
glutamate receptor interacting protein 2b |
| chr2_-_36819624 | 0.07 |
ENSDART00000140844
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr3_+_39664256 | 0.06 |
ENSDART00000153866
|
epn2
|
epsin 2 |
| chr14_-_30945515 | 0.06 |
ENSDART00000161540
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr14_-_12071447 | 0.06 |
ENSDART00000166116
|
tmsb1
|
thymosin beta 1 |
| chr9_-_30808073 | 0.05 |
ENSDART00000146300
|
klf5b
|
Kruppel-like factor 5b |
| chr24_-_36727922 | 0.05 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr5_+_3501859 | 0.05 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr5_-_25582721 | 0.05 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr12_-_19862912 | 0.04 |
ENSDART00000145788
|
shisa9a
|
shisa family member 9a |
| chr5_+_32162684 | 0.04 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr5_-_25583125 | 0.03 |
ENSDART00000031665
ENSDART00000145353 |
anxa1a
|
annexin A1a |
| chr21_-_43949208 | 0.03 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr9_-_6927587 | 0.03 |
ENSDART00000059092
|
tmem182a
|
transmembrane protein 182a |
| chr9_+_9502610 | 0.03 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr10_-_29733194 | 0.03 |
ENSDART00000149252
|
si:ch73-261i21.2
|
si:ch73-261i21.2 |
| chr8_-_31716302 | 0.02 |
ENSDART00000061832
|
si:dkey-46a10.3
|
si:dkey-46a10.3 |
| chr23_+_11285662 | 0.02 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr4_-_17391091 | 0.02 |
ENSDART00000056002
|
th2
|
tyrosine hydroxylase 2 |
| chr7_+_15736230 | 0.02 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr21_+_1647990 | 0.02 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr16_+_44298902 | 0.01 |
ENSDART00000114795
|
dpys
|
dihydropyrimidinase |
| chr22_+_15336752 | 0.01 |
ENSDART00000139070
|
sult3st2
|
sulfotransferase family 3, cytosolic sulfotransferase 2 |
| chr5_-_58840971 | 0.01 |
ENSDART00000050932
|
tmem136b
|
transmembrane protein 136b |
| chr12_+_35119762 | 0.01 |
ENSDART00000085774
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr21_-_40174647 | 0.00 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr15_+_20543770 | 0.00 |
ENSDART00000092357
|
sgsm2
|
small G protein signaling modulator 2 |
| chr7_+_23580082 | 0.00 |
ENSDART00000183245
|
si:dkey-172j4.3
|
si:dkey-172j4.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of cebpd
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.3 | 1.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.3 | 0.8 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 0.2 | 0.7 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.2 | 1.6 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.2 | 1.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 1.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.7 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 0.3 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.5 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.3 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) mediolateral intercalation(GO:0060031) |
| 0.1 | 1.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.4 | GO:0045041 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.4 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 1.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.3 | GO:0003418 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.1 | 0.4 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.6 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 3.8 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 1.0 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.3 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.6 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.7 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 1.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.6 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.2 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.7 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 1.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.6 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 1.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.7 | 2.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.2 | 1.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 1.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.6 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0030669 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.3 | 1.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.2 | 1.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 0.7 | GO:0072591 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.2 | 0.9 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.2 | 1.2 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.7 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 1.0 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 1.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.0 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 3.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.3 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.6 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 2.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.6 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.6 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.8 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |