Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
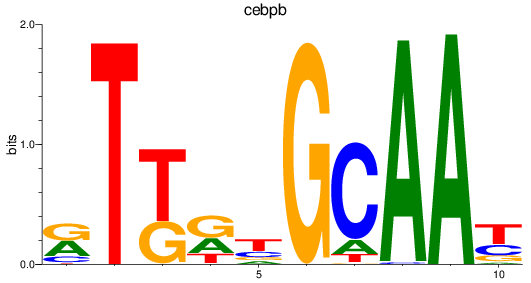
Results for cebpb
Z-value: 0.43
Transcription factors associated with cebpb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebpb
|
ENSDARG00000042725 | CCAAT enhancer binding protein beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cebpb | dr11_v1_chr8_-_28449782_28449782 | -0.50 | 3.3e-02 | Click! |
Activity profile of cebpb motif
Sorted Z-values of cebpb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_46020508 | 1.44 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr5_+_22974019 | 1.27 |
ENSDART00000147157
ENSDART00000020434 |
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr1_-_9486214 | 1.25 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr1_-_9485939 | 1.25 |
ENSDART00000157814
|
micall2b
|
mical-like 2b |
| chr13_+_40437550 | 1.23 |
ENSDART00000057090
ENSDART00000167859 |
got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr9_-_53062083 | 1.19 |
ENSDART00000122155
|
zmp:0000000936
|
zmp:0000000936 |
| chr7_+_46019780 | 1.18 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr20_-_32045057 | 0.99 |
ENSDART00000152970
ENSDART00000034248 |
rab32a
|
RAB32a, member RAS oncogene family |
| chr19_-_25113660 | 0.99 |
ENSDART00000035538
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr10_+_15255012 | 0.96 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr10_+_15255198 | 0.93 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr9_+_44304980 | 0.87 |
ENSDART00000147990
|
ssfa2
|
sperm specific antigen 2 |
| chr3_+_36617024 | 0.84 |
ENSDART00000189957
|
pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr12_+_30789611 | 0.81 |
ENSDART00000181501
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr9_-_32300783 | 0.79 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr12_+_4920451 | 0.78 |
ENSDART00000171525
ENSDART00000159986 |
plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr3_-_30885250 | 0.78 |
ENSDART00000109104
|
kmt5c
|
lysine methyltransferase 5C |
| chr3_-_26190804 | 0.77 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr9_-_32300611 | 0.75 |
ENSDART00000127938
|
hspd1
|
heat shock 60 protein 1 |
| chr5_-_15283509 | 0.74 |
ENSDART00000052712
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr16_+_47207691 | 0.73 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr11_-_34577034 | 0.73 |
ENSDART00000133302
ENSDART00000184367 |
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr6_+_23810529 | 0.69 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr7_+_27691647 | 0.68 |
ENSDART00000079091
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr5_-_69948099 | 0.65 |
ENSDART00000034639
ENSDART00000191111 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr17_+_12942021 | 0.64 |
ENSDART00000192514
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr14_-_5642371 | 0.62 |
ENSDART00000183859
ENSDART00000054876 |
npm1b
|
nucleophosmin 1b |
| chr15_+_24676905 | 0.62 |
ENSDART00000078014
ENSDART00000143137 |
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr18_-_37252036 | 0.62 |
ENSDART00000132230
|
six5
|
SIX homeobox 5 |
| chr3_-_32873641 | 0.61 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr17_-_12058171 | 0.60 |
ENSDART00000105236
|
smyd3
|
SET and MYND domain containing 3 |
| chr16_+_25259313 | 0.60 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr12_+_46869271 | 0.59 |
ENSDART00000166560
|
hpdl
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr23_-_29394505 | 0.59 |
ENSDART00000017728
|
pgd
|
phosphogluconate dehydrogenase |
| chr8_+_23827571 | 0.58 |
ENSDART00000040362
|
mapk14a
|
mitogen-activated protein kinase 14a |
| chr20_+_28266892 | 0.58 |
ENSDART00000103330
|
chac1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr14_-_7306983 | 0.57 |
ENSDART00000158914
|
si:ch211-51f19.1
|
si:ch211-51f19.1 |
| chr21_+_17051478 | 0.57 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr18_-_20444296 | 0.55 |
ENSDART00000132993
|
kif23
|
kinesin family member 23 |
| chr11_+_2710530 | 0.55 |
ENSDART00000132768
ENSDART00000030921 ENSDART00000040147 |
mapk14b
|
mitogen-activated protein kinase 14b |
| chr6_+_8314451 | 0.55 |
ENSDART00000147793
ENSDART00000183688 |
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr8_+_21229718 | 0.55 |
ENSDART00000100222
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr3_+_32663865 | 0.54 |
ENSDART00000190240
|
si:dkey-16l2.16
|
si:dkey-16l2.16 |
| chr14_-_897874 | 0.54 |
ENSDART00000167395
|
rgs14a
|
regulator of G protein signaling 14a |
| chr9_-_27398369 | 0.53 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr18_+_17493859 | 0.53 |
ENSDART00000090754
|
si:dkey-102f14.5
|
si:dkey-102f14.5 |
| chr15_+_24737599 | 0.52 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr17_+_12942634 | 0.52 |
ENSDART00000016597
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr4_-_4261673 | 0.51 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr8_+_26874924 | 0.51 |
ENSDART00000141794
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr23_+_38957472 | 0.50 |
ENSDART00000193836
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr20_+_43113465 | 0.50 |
ENSDART00000004842
|
dusp23a
|
dual specificity phosphatase 23a |
| chr6_-_39631164 | 0.50 |
ENSDART00000104042
ENSDART00000136076 |
atf7b
|
activating transcription factor 7b |
| chr15_-_24960730 | 0.50 |
ENSDART00000109990
ENSDART00000186706 |
abhd15a
|
abhydrolase domain containing 15a |
| chr6_+_8315050 | 0.50 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr9_-_30264415 | 0.49 |
ENSDART00000060150
|
mid1ip1a
|
MID1 interacting protein 1a |
| chr23_+_38957738 | 0.49 |
ENSDART00000193480
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr24_+_41989108 | 0.49 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr19_-_11014641 | 0.49 |
ENSDART00000183745
|
tpm3
|
tropomyosin 3 |
| chr12_+_13205955 | 0.48 |
ENSDART00000092906
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr6_-_41138854 | 0.48 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr9_-_13871935 | 0.48 |
ENSDART00000146597
|
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr14_+_25505468 | 0.47 |
ENSDART00000079016
|
thoc3
|
THO complex 3 |
| chr24_+_9881219 | 0.47 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr24_+_26134029 | 0.46 |
ENSDART00000185134
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr22_-_17677947 | 0.46 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr20_+_47491247 | 0.45 |
ENSDART00000113412
|
lin28b
|
lin-28 homolog B (C. elegans) |
| chr6_+_44197348 | 0.45 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr6_+_11397269 | 0.44 |
ENSDART00000114260
|
senp2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr11_+_31323746 | 0.44 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr8_+_42917515 | 0.44 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr17_+_33158350 | 0.43 |
ENSDART00000104476
|
snx9a
|
sorting nexin 9a |
| chr16_-_30655980 | 0.42 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr14_+_31493306 | 0.42 |
ENSDART00000138341
|
phf6
|
PHD finger protein 6 |
| chr8_-_26961779 | 0.42 |
ENSDART00000099214
|
slc16a1b
|
solute carrier family 16 (monocarboxylate transporter), member 1b |
| chr6_+_44197099 | 0.42 |
ENSDART00000124168
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr22_-_10397600 | 0.41 |
ENSDART00000181964
ENSDART00000142886 |
nisch
|
nischarin |
| chr9_+_33154841 | 0.41 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr14_+_31493119 | 0.41 |
ENSDART00000006463
|
phf6
|
PHD finger protein 6 |
| chr13_+_30035253 | 0.41 |
ENSDART00000181303
ENSDART00000057525 ENSDART00000136622 |
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr9_-_41090048 | 0.40 |
ENSDART00000131681
ENSDART00000182552 |
asnsd1
|
asparagine synthetase domain containing 1 |
| chr8_+_28724692 | 0.39 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr6_+_27090800 | 0.39 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr11_+_24314148 | 0.39 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr11_-_44931962 | 0.38 |
ENSDART00000170345
|
pfklb
|
phosphofructokinase, liver b |
| chr18_-_15551360 | 0.38 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr25_+_15997957 | 0.38 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr21_+_4509483 | 0.37 |
ENSDART00000025612
|
phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr17_+_37310663 | 0.37 |
ENSDART00000157122
|
elmsan1b
|
ELM2 and Myb/SANT-like domain containing 1b |
| chr16_-_42770064 | 0.37 |
ENSDART00000112879
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr17_+_5623514 | 0.37 |
ENSDART00000171220
ENSDART00000176083 |
CU571310.1
|
|
| chr1_+_54737353 | 0.37 |
ENSDART00000130675
ENSDART00000162075 |
pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr4_-_19693978 | 0.36 |
ENSDART00000100974
ENSDART00000040405 |
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr6_-_10728057 | 0.35 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr5_-_67895656 | 0.35 |
ENSDART00000158917
|
abhd10a
|
abhydrolase domain containing 10a |
| chr9_-_34500197 | 0.35 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr12_-_32073611 | 0.35 |
ENSDART00000153369
ENSDART00000086531 |
cox11
|
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr5_-_6745442 | 0.35 |
ENSDART00000157402
ENSDART00000128684 ENSDART00000168698 |
ostf1
|
osteoclast stimulating factor 1 |
| chr3_-_40836081 | 0.34 |
ENSDART00000143135
|
wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr14_+_3507326 | 0.34 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr1_+_38142354 | 0.34 |
ENSDART00000179352
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr12_-_17199381 | 0.34 |
ENSDART00000193292
|
lipf
|
lipase, gastric |
| chr16_+_33143503 | 0.33 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr3_-_40275096 | 0.33 |
ENSDART00000141578
|
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr2_-_54387550 | 0.33 |
ENSDART00000097388
|
napgb
|
N-ethylmaleimide-sensitive factor attachment protein, gamma b |
| chr15_+_35933094 | 0.33 |
ENSDART00000019976
|
rhbdd1
|
rhomboid domain containing 1 |
| chr5_-_32336613 | 0.32 |
ENSDART00000139732
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr9_+_24065855 | 0.32 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr2_+_36616830 | 0.32 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr15_-_1885247 | 0.32 |
ENSDART00000149703
|
porb
|
P450 (cytochrome) oxidoreductase b |
| chr25_-_32869794 | 0.32 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr11_+_23933016 | 0.31 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr20_-_33675676 | 0.31 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr23_-_25135046 | 0.31 |
ENSDART00000184844
ENSDART00000103989 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr16_-_27138478 | 0.31 |
ENSDART00000147438
|
tmem245
|
transmembrane protein 245 |
| chr14_+_48862987 | 0.31 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr2_-_49860723 | 0.31 |
ENSDART00000083690
|
blvra
|
biliverdin reductase A |
| chr12_-_13205572 | 0.31 |
ENSDART00000152670
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr10_+_45248326 | 0.30 |
ENSDART00000159797
|
zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr7_+_38089650 | 0.30 |
ENSDART00000052365
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr24_-_20208474 | 0.30 |
ENSDART00000139329
|
cry-dash
|
cryptochrome DASH |
| chr7_+_16033923 | 0.29 |
ENSDART00000161669
ENSDART00000114062 |
immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr14_-_15990361 | 0.29 |
ENSDART00000168075
|
trim105
|
tripartite motif containing 105 |
| chr5_+_8196264 | 0.29 |
ENSDART00000174564
ENSDART00000161261 |
lmbrd2a
|
LMBR1 domain containing 2a |
| chr8_+_26007988 | 0.29 |
ENSDART00000193948
ENSDART00000058100 |
xpc
|
xeroderma pigmentosum, complementation group C |
| chr1_-_19215336 | 0.28 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr6_-_10708960 | 0.28 |
ENSDART00000157704
|
si:dkey-34m19.3
|
si:dkey-34m19.3 |
| chr13_+_30163515 | 0.28 |
ENSDART00000040926
|
eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr3_+_19685873 | 0.28 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr22_-_11517377 | 0.28 |
ENSDART00000193885
|
slc26a11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr6_+_22068589 | 0.28 |
ENSDART00000151205
|
aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr2_-_37277626 | 0.27 |
ENSDART00000135340
|
nadkb
|
NAD kinase b |
| chr16_-_9830451 | 0.27 |
ENSDART00000148528
|
grhl2a
|
grainyhead-like transcription factor 2a |
| chr2_-_36819624 | 0.27 |
ENSDART00000140844
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr12_-_22238004 | 0.27 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr6_+_45692026 | 0.26 |
ENSDART00000164759
|
cntn4
|
contactin 4 |
| chr20_+_39223235 | 0.26 |
ENSDART00000132132
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr8_-_35960987 | 0.26 |
ENSDART00000160503
|
slc15a4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr9_-_5318873 | 0.26 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr7_+_38090515 | 0.26 |
ENSDART00000131387
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr7_-_29723761 | 0.25 |
ENSDART00000173560
|
vps13c
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr5_+_44346691 | 0.25 |
ENSDART00000034523
|
tars
|
threonyl-tRNA synthetase |
| chr20_+_38458084 | 0.25 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr18_-_21047007 | 0.25 |
ENSDART00000162702
|
igf1ra
|
insulin-like growth factor 1a receptor |
| chr22_-_16377666 | 0.24 |
ENSDART00000161878
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr14_-_30905288 | 0.24 |
ENSDART00000173449
ENSDART00000173451 |
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr6_-_18751696 | 0.24 |
ENSDART00000171537
|
tnrc6c2
|
trinucleotide repeat containing 6C2 |
| chr12_-_28349026 | 0.24 |
ENSDART00000183768
ENSDART00000152998 |
zgc:195081
|
zgc:195081 |
| chr9_+_8968702 | 0.24 |
ENSDART00000008490
|
ube2al
|
ubiquitin conjugating enzyme E2 A, like |
| chr11_+_30253968 | 0.24 |
ENSDART00000157272
ENSDART00000003475 |
ppef1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr2_+_11029138 | 0.23 |
ENSDART00000138737
ENSDART00000081058 ENSDART00000153662 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr1_+_2260407 | 0.23 |
ENSDART00000058876
|
kpnb3
|
karyopherin (importin) beta 3 |
| chr9_-_9732212 | 0.23 |
ENSDART00000092884
|
lrrc58b
|
leucine rich repeat containing 58b |
| chr14_+_7902374 | 0.23 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr11_-_2250767 | 0.22 |
ENSDART00000018131
|
hnrnpa1a
|
heterogeneous nuclear ribonucleoprotein A1a |
| chr24_-_31123365 | 0.22 |
ENSDART00000182947
|
tmem56a
|
transmembrane protein 56a |
| chr13_+_41917606 | 0.22 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr17_-_17759138 | 0.22 |
ENSDART00000157128
ENSDART00000123845 |
adck1
|
aarF domain containing kinase 1 |
| chr1_-_22412042 | 0.21 |
ENSDART00000074678
|
chrnb3a
|
cholinergic receptor, nicotinic, beta polypeptide 3a |
| chr11_-_17803071 | 0.21 |
ENSDART00000080752
ENSDART00000153801 |
uba3
|
ubiquitin-like modifier activating enzyme 3 |
| chr3_+_33440615 | 0.21 |
ENSDART00000146005
|
gtpbp1
|
GTP binding protein 1 |
| chr12_+_36109507 | 0.21 |
ENSDART00000175409
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr23_+_39413163 | 0.21 |
ENSDART00000184254
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr20_+_37794633 | 0.21 |
ENSDART00000022060
|
atf3
|
activating transcription factor 3 |
| chr24_+_26134209 | 0.20 |
ENSDART00000038824
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr5_-_8711157 | 0.20 |
ENSDART00000062957
|
paip1
|
poly(A) binding protein interacting protein 1 |
| chr7_+_21787507 | 0.20 |
ENSDART00000100936
|
tmem88b
|
transmembrane protein 88 b |
| chr18_-_26797723 | 0.20 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr10_-_35613752 | 0.19 |
ENSDART00000158537
|
smg6
|
SMG6 nonsense mediated mRNA decay factor |
| chr8_+_16990120 | 0.19 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr6_+_42475730 | 0.19 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr22_+_25931782 | 0.19 |
ENSDART00000157842
|
dnaja3b
|
DnaJ (Hsp40) homolog, subfamily A, member 3B |
| chr14_+_16345003 | 0.19 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr22_-_16377960 | 0.18 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr3_-_26978793 | 0.18 |
ENSDART00000155396
|
nubp1
|
nucleotide binding protein 1 (MinD homolog, E. coli) |
| chr7_+_11543999 | 0.18 |
ENSDART00000173676
|
il16
|
interleukin 16 |
| chr3_+_3641429 | 0.18 |
ENSDART00000092393
|
plbd1
|
phospholipase B domain containing 1 |
| chr6_-_25371196 | 0.17 |
ENSDART00000187291
|
PKN2 (1 of many)
|
zgc:153916 |
| chr15_-_31419805 | 0.17 |
ENSDART00000060111
|
or111-11
|
odorant receptor, family D, subfamily 111, member 11 |
| chr7_+_24081167 | 0.17 |
ENSDART00000141749
|
acin1b
|
apoptotic chromatin condensation inducer 1b |
| chr1_-_18585046 | 0.16 |
ENSDART00000147228
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr14_+_46003973 | 0.16 |
ENSDART00000145241
ENSDART00000141545 |
slu7
|
SLU7 homolog, splicing factor |
| chr18_+_15644559 | 0.16 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr21_-_30160411 | 0.16 |
ENSDART00000126621
|
pfdn1
|
prefoldin subunit 1 |
| chr24_-_32582378 | 0.16 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr7_-_60831082 | 0.16 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr9_+_22375331 | 0.16 |
ENSDART00000090907
|
dgkg
|
diacylglycerol kinase, gamma |
| chr21_-_15929041 | 0.15 |
ENSDART00000080693
|
lhx5
|
LIM homeobox 5 |
| chr12_-_19862912 | 0.15 |
ENSDART00000145788
|
shisa9a
|
shisa family member 9a |
| chr24_-_38644937 | 0.15 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr12_+_30368145 | 0.15 |
ENSDART00000153454
|
ccdc186
|
si:ch211-225b10.4 |
| chr11_-_29082175 | 0.15 |
ENSDART00000123245
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr24_-_39771667 | 0.14 |
ENSDART00000181867
|
GFOD1
|
si:ch211-276f18.2 |
| chr21_-_11646878 | 0.14 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr9_+_22375779 | 0.14 |
ENSDART00000183956
|
dgkg
|
diacylglycerol kinase, gamma |
| chr19_-_15192840 | 0.14 |
ENSDART00000151337
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr15_+_45994123 | 0.14 |
ENSDART00000124704
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr21_+_29227224 | 0.14 |
ENSDART00000013641
|
adra1ba
|
adrenoceptor alpha 1Ba |
| chr8_-_23599096 | 0.14 |
ENSDART00000183096
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr4_+_16784553 | 0.14 |
ENSDART00000134054
|
si:dkey-13i19.8
|
si:dkey-13i19.8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of cebpb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0045041 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.3 | 1.2 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.3 | 0.8 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.2 | 1.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 0.6 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.4 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 1.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.7 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.1 | 0.3 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.3 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.3 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 0.4 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.7 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.3 | GO:0009397 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.9 | GO:1903318 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.1 | 0.1 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.1 | 0.4 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.2 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.6 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.6 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.3 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 2.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.6 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 2.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.5 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.3 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.4 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.7 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.3 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 0.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.4 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 2.0 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.8 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.4 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.0 | 0.1 | GO:0003321 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.4 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.6 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.0 | GO:0060959 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.2 | GO:1900151 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.5 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.5 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 1.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.8 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.6 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.3 | 0.9 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 1.8 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.1 | 0.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.5 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.3 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.3 | 1.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.3 | 1.0 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.3 | 0.8 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.2 | 1.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.2 | 0.8 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.2 | 0.6 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.2 | 0.5 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.2 | 0.8 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.6 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.3 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.1 | 0.3 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.1 | 0.4 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.7 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.3 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 0.5 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.1 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 2.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 1.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 1.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0032896 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 1.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 1.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |