Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
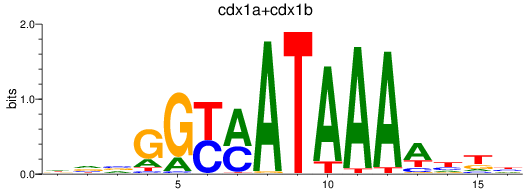
Results for cdx1a+cdx1b
Z-value: 0.87
Transcription factors associated with cdx1a+cdx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cdx1a
|
ENSDARG00000114554 | caudal type homeobox 1a |
|
cdx1b
|
ENSDARG00000116139 | caudal type homeobox 1 b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cdx1a | dr11_v1_chr14_+_3287740_3287740 | 0.37 | 1.3e-01 | Click! |
| cdx1b | dr11_v1_chr21_-_43992027_43992027 | 0.04 | 8.7e-01 | Click! |
Activity profile of cdx1a+cdx1b motif
Sorted Z-values of cdx1a+cdx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_53468160 | 2.04 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr3_-_53091946 | 2.04 |
ENSDART00000187297
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr1_-_13989643 | 1.88 |
ENSDART00000191046
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr15_-_28596507 | 1.48 |
ENSDART00000156800
|
si:ch211-225b7.5
|
si:ch211-225b7.5 |
| chr3_+_49074008 | 1.48 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr20_+_52546186 | 1.44 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr24_-_9997948 | 1.26 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr10_-_21545091 | 1.15 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr12_+_3871452 | 1.11 |
ENSDART00000066546
|
nif3l1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr9_-_710896 | 1.11 |
ENSDART00000180478
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr11_+_25257022 | 1.09 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr20_-_43741159 | 1.09 |
ENSDART00000192621
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr18_-_14691727 | 1.08 |
ENSDART00000010129
|
pdf
|
peptide deformylase, mitochondrial |
| chr21_-_11855828 | 1.03 |
ENSDART00000081666
|
ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr10_+_44700103 | 1.00 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr15_+_32405959 | 0.95 |
ENSDART00000177269
|
si:ch211-162k9.6
|
si:ch211-162k9.6 |
| chr9_+_3282369 | 0.95 |
ENSDART00000044128
|
hat1
|
histone acetyltransferase 1 |
| chr10_+_40321067 | 0.93 |
ENSDART00000109816
|
gltpb
|
glycolipid transfer protein b |
| chr5_-_1963498 | 0.91 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr6_+_120181 | 0.86 |
ENSDART00000151209
ENSDART00000185930 |
cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr19_+_3840955 | 0.86 |
ENSDART00000172305
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr20_+_26880668 | 0.86 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr5_+_58687541 | 0.85 |
ENSDART00000083015
ENSDART00000181902 |
ccdc84
|
coiled-coil domain containing 84 |
| chr14_-_45604667 | 0.84 |
ENSDART00000167016
|
si:ch211-276i12.9
|
si:ch211-276i12.9 |
| chr6_-_6976096 | 0.81 |
ENSDART00000151822
ENSDART00000039443 ENSDART00000177960 |
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr2_-_5728843 | 0.81 |
ENSDART00000014020
|
sst2
|
somatostatin 2 |
| chr24_-_10006158 | 0.78 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr5_-_9073433 | 0.78 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr10_+_44699734 | 0.78 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr1_+_8508753 | 0.76 |
ENSDART00000091683
|
alkbh5
|
alkB homolog 5, RNA demethylase |
| chr9_+_51891 | 0.76 |
ENSDART00000163529
|
zgc:158316
|
zgc:158316 |
| chr5_+_70155935 | 0.73 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr20_-_13140309 | 0.73 |
ENSDART00000020703
ENSDART00000188594 |
ints7
|
integrator complex subunit 7 |
| chr25_+_16080181 | 0.73 |
ENSDART00000061753
|
far1
|
fatty acyl CoA reductase 1 |
| chr12_+_27022517 | 0.72 |
ENSDART00000152975
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr24_-_10014512 | 0.71 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr3_-_29910547 | 0.69 |
ENSDART00000151501
|
RUNDC1
|
si:dkey-151m15.5 |
| chr22_+_2512154 | 0.69 |
ENSDART00000097363
|
zgc:173726
|
zgc:173726 |
| chr23_+_7379728 | 0.67 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr16_+_43344475 | 0.67 |
ENSDART00000085282
|
mrpl32
|
mitochondrial ribosomal protein L32 |
| chr24_-_24114217 | 0.67 |
ENSDART00000160578
|
zgc:112982
|
zgc:112982 |
| chr15_-_4580763 | 0.66 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr7_-_48263516 | 0.65 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr15_+_32423801 | 0.65 |
ENSDART00000165591
|
si:dkey-285b23.3
|
si:dkey-285b23.3 |
| chr10_-_15340362 | 0.65 |
ENSDART00000148119
ENSDART00000127277 ENSDART00000154037 ENSDART00000189109 |
pum3
|
pumilio RNA-binding family member 3 |
| chr2_-_27975530 | 0.64 |
ENSDART00000020367
|
zgc:56556
|
zgc:56556 |
| chr3_-_15999501 | 0.62 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr7_+_52154215 | 0.61 |
ENSDART00000098712
|
TMEM208
|
zgc:77041 |
| chr21_-_15929041 | 0.61 |
ENSDART00000080693
|
lhx5
|
LIM homeobox 5 |
| chr12_-_13205854 | 0.59 |
ENSDART00000077829
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr21_+_20396858 | 0.59 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr11_-_25538341 | 0.57 |
ENSDART00000171560
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr24_-_6158933 | 0.57 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr5_+_64856666 | 0.56 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr21_-_41838284 | 0.56 |
ENSDART00000141067
|
cct6a
|
chaperonin containing TCP1, subunit 6A (zeta 1) |
| chr15_+_42285643 | 0.54 |
ENSDART00000152731
|
scaf4b
|
SR-related CTD-associated factor 4b |
| chr23_+_28092083 | 0.53 |
ENSDART00000053958
|
c1galt1a
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1a |
| chr18_-_44908479 | 0.52 |
ENSDART00000169636
|
si:ch211-71n6.4
|
si:ch211-71n6.4 |
| chr24_+_26134029 | 0.52 |
ENSDART00000185134
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr1_+_41466011 | 0.52 |
ENSDART00000135280
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr11_+_44622472 | 0.51 |
ENSDART00000159068
ENSDART00000166323 ENSDART00000187753 |
rbm34
|
RNA binding motif protein 34 |
| chr1_+_34496855 | 0.50 |
ENSDART00000012873
|
klf12a
|
Kruppel-like factor 12a |
| chr1_-_9249943 | 0.49 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr14_-_9056066 | 0.49 |
ENSDART00000139669
ENSDART00000138758 ENSDART00000041099 |
sybl1
|
synaptobrevin-like 1 |
| chr2_+_26682913 | 0.48 |
ENSDART00000010683
ENSDART00000131411 ENSDART00000137933 |
impa1
|
inositol monophosphatase 1 |
| chr15_-_23908779 | 0.48 |
ENSDART00000088808
|
usp32
|
ubiquitin specific peptidase 32 |
| chr1_+_41465771 | 0.48 |
ENSDART00000010211
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr14_-_4121052 | 0.48 |
ENSDART00000167074
|
irf2
|
interferon regulatory factor 2 |
| chr24_+_30392834 | 0.48 |
ENSDART00000162555
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr5_-_69499486 | 0.48 |
ENSDART00000023983
ENSDART00000180293 |
psat1
|
phosphoserine aminotransferase 1 |
| chr14_-_41272034 | 0.47 |
ENSDART00000191709
ENSDART00000074438 |
cenpi
|
centromere protein I |
| chr21_+_41837610 | 0.46 |
ENSDART00000157974
|
rnf14
|
ring finger protein 14 |
| chr5_-_67750907 | 0.45 |
ENSDART00000172097
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr25_-_35047838 | 0.45 |
ENSDART00000155060
|
CU302436.3
|
|
| chr14_-_15171435 | 0.45 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr7_+_73827805 | 0.44 |
ENSDART00000109316
|
zgc:173587
|
zgc:173587 |
| chr23_+_44349252 | 0.44 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr24_+_39105051 | 0.42 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr3_-_33437796 | 0.41 |
ENSDART00000075499
|
si:dkey-283b1.7
|
si:dkey-283b1.7 |
| chr10_+_27068251 | 0.41 |
ENSDART00000012717
|
ehd1b
|
EH-domain containing 1b |
| chr12_-_48467733 | 0.41 |
ENSDART00000153126
ENSDART00000152895 ENSDART00000014190 |
sec31b
|
SEC31 homolog B, COPII coat complex component |
| chr13_-_5252559 | 0.41 |
ENSDART00000181652
|
si:dkey-78p8.1
|
si:dkey-78p8.1 |
| chr14_+_15495088 | 0.40 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr2_+_2223837 | 0.40 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr23_-_24699400 | 0.39 |
ENSDART00000104031
|
sdf4
|
stromal cell derived factor 4 |
| chr19_+_19652439 | 0.39 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr25_-_20258508 | 0.38 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr2_+_52847049 | 0.37 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr7_+_66822229 | 0.37 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr13_-_15793585 | 0.36 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2 associated athanogene 5 |
| chr13_-_36034582 | 0.36 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr1_-_43892349 | 0.34 |
ENSDART00000148416
|
tacr3a
|
tachykinin receptor 3a |
| chr14_+_35369979 | 0.32 |
ENSDART00000144702
ENSDART00000169712 |
clint1a
|
clathrin interactor 1a |
| chr15_-_23908605 | 0.32 |
ENSDART00000185578
|
usp32
|
ubiquitin specific peptidase 32 |
| chr16_+_42481447 | 0.31 |
ENSDART00000037401
|
herpud2
|
HERPUD family member 2 |
| chr16_-_1757521 | 0.31 |
ENSDART00000124660
|
ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr12_-_2993095 | 0.31 |
ENSDART00000152316
|
si:dkey-202c14.3
|
si:dkey-202c14.3 |
| chr25_-_27633115 | 0.31 |
ENSDART00000131539
|
hyal6
|
hyaluronoglucosaminidase 6 |
| chr22_+_34616151 | 0.31 |
ENSDART00000155399
ENSDART00000104705 |
si:ch1073-214b20.2
|
si:ch1073-214b20.2 |
| chr14_-_4120636 | 0.31 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr21_+_26720803 | 0.31 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr5_-_31856681 | 0.31 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr12_-_45238759 | 0.31 |
ENSDART00000154859
|
trim65
|
tripartite motif containing 65 |
| chr13_-_25719628 | 0.30 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr10_-_3332362 | 0.30 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr23_+_309099 | 0.30 |
ENSDART00000134556
ENSDART00000055152 |
taf11
|
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr9_-_16853462 | 0.29 |
ENSDART00000160273
|
CT573248.2
|
|
| chr10_+_26926654 | 0.29 |
ENSDART00000078980
ENSDART00000100289 |
rab1bb
|
RAB1B, member RAS oncogene family b |
| chr10_+_26612321 | 0.29 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr23_-_39751234 | 0.28 |
ENSDART00000160853
|
minos1
|
mitochondrial inner membrane organizing system 1 |
| chr19_+_1873059 | 0.28 |
ENSDART00000145246
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr11_-_44622240 | 0.28 |
ENSDART00000172258
ENSDART00000173340 |
tbce
|
tubulin folding cofactor E |
| chr12_+_23912074 | 0.28 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr2_+_42592572 | 0.27 |
ENSDART00000179722
|
march11
|
membrane-associated ring finger (C3HC4) 11 |
| chr7_-_44672095 | 0.27 |
ENSDART00000073745
|
cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr10_-_43721530 | 0.27 |
ENSDART00000025366
|
cetn3
|
centrin 3 |
| chr6_-_40697585 | 0.27 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr20_-_14924858 | 0.27 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
| chr17_-_43666166 | 0.27 |
ENSDART00000077990
|
egr2a
|
early growth response 2a |
| chr8_+_37755099 | 0.26 |
ENSDART00000075708
ENSDART00000140966 |
got1l1
|
glutamic-oxaloacetic transaminase 1 like 1 |
| chr24_+_17270129 | 0.26 |
ENSDART00000186729
|
spag6
|
sperm associated antigen 6 |
| chr6_+_49551614 | 0.26 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr8_+_30709685 | 0.26 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr5_-_16351306 | 0.26 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr25_+_17847054 | 0.26 |
ENSDART00000067290
|
PYURF
|
zgc:162634 |
| chr10_-_10864331 | 0.25 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr20_-_14925281 | 0.25 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr4_-_8795250 | 0.24 |
ENSDART00000169823
|
pnpla3
|
patatin-like phospholipase domain containing 3 |
| chr3_-_35554809 | 0.24 |
ENSDART00000010944
|
dctn5
|
dynactin 5 |
| chr1_+_30103297 | 0.24 |
ENSDART00000146783
|
vps8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr10_-_31563049 | 0.23 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr23_-_18030399 | 0.23 |
ENSDART00000136967
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr18_+_14684115 | 0.23 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr5_-_8406192 | 0.22 |
ENSDART00000159718
|
spef2
|
sperm flagellar 2 |
| chr10_-_24371312 | 0.22 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr16_+_42471455 | 0.22 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr11_-_44622478 | 0.21 |
ENSDART00000171837
|
tbce
|
tubulin folding cofactor E |
| chr24_+_32592748 | 0.21 |
ENSDART00000188256
|
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr3_-_19200571 | 0.21 |
ENSDART00000131503
ENSDART00000012335 |
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr14_-_12020653 | 0.20 |
ENSDART00000106654
|
znf711
|
zinc finger protein 711 |
| chr15_+_29276508 | 0.20 |
ENSDART00000170537
ENSDART00000126559 |
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr7_-_41693004 | 0.20 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr7_+_18989241 | 0.20 |
ENSDART00000053213
|
zgc:194252
|
zgc:194252 |
| chr5_-_12713920 | 0.19 |
ENSDART00000099749
|
CLDN22
|
si:dkey-98f17.3 |
| chr6_+_40629066 | 0.19 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr10_-_35410518 | 0.19 |
ENSDART00000048430
|
gabrr3a
|
gamma-aminobutyric acid (GABA) A receptor, rho 3a |
| chr8_-_38159805 | 0.18 |
ENSDART00000112331
ENSDART00000180006 |
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr5_-_9625459 | 0.18 |
ENSDART00000143347
|
sh2b3
|
SH2B adaptor protein 3 |
| chr16_-_24561354 | 0.18 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr11_-_36051004 | 0.17 |
ENSDART00000025033
|
gpx1a
|
glutathione peroxidase 1a |
| chr11_-_18323059 | 0.17 |
ENSDART00000182590
|
SFMBT1
|
Scm like with four mbt domains 1 |
| chr11_-_21303946 | 0.17 |
ENSDART00000185786
|
RASSF5
|
si:dkey-85p17.3 |
| chr10_-_34889053 | 0.17 |
ENSDART00000136966
|
ccdc169
|
coiled-coil domain containing 169 |
| chr7_+_22823889 | 0.17 |
ENSDART00000127467
ENSDART00000148576 ENSDART00000149993 |
pygmb
|
phosphorylase, glycogen, muscle b |
| chr8_-_53169445 | 0.16 |
ENSDART00000144493
|
rbsn
|
rabenosyn, RAB effector |
| chr24_+_26402110 | 0.16 |
ENSDART00000133684
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr10_-_31562695 | 0.16 |
ENSDART00000186456
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr13_+_4405282 | 0.16 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr10_-_29903165 | 0.16 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr25_-_19433244 | 0.16 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr22_+_34615591 | 0.16 |
ENSDART00000189563
|
si:ch1073-214b20.2
|
si:ch1073-214b20.2 |
| chr11_+_12052791 | 0.16 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr6_-_12588044 | 0.16 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr11_+_6116503 | 0.16 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr5_-_23715861 | 0.16 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr15_-_3956906 | 0.16 |
ENSDART00000171202
|
clrn1
|
clarin 1 |
| chr10_-_44924289 | 0.16 |
ENSDART00000171267
|
tuba7l
|
tubulin, alpha 7 like |
| chr12_+_18744610 | 0.16 |
ENSDART00000153456
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr25_+_29161609 | 0.16 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr25_+_4750972 | 0.16 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr5_-_31857345 | 0.15 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr8_+_18615938 | 0.15 |
ENSDART00000089161
|
si:ch211-232d19.4
|
si:ch211-232d19.4 |
| chr25_-_18953322 | 0.15 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr7_+_50766094 | 0.15 |
ENSDART00000165037
|
si:ch73-380l10.2
|
si:ch73-380l10.2 |
| chr21_+_11969603 | 0.15 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr24_-_40009446 | 0.15 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr16_+_13993285 | 0.15 |
ENSDART00000139130
ENSDART00000130353 |
si:dkey-85k15.7
fdps
|
si:dkey-85k15.7 farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr2_+_5927255 | 0.15 |
ENSDART00000152866
|
si:ch211-168b3.2
|
si:ch211-168b3.2 |
| chr23_-_39959784 | 0.14 |
ENSDART00000115046
|
xcr1a.1
|
chemokine (C motif) receptor 1a, duplicate 1 |
| chr14_-_31060082 | 0.14 |
ENSDART00000111601
ENSDART00000161113 |
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr5_-_30984271 | 0.14 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr23_-_39959173 | 0.14 |
ENSDART00000077122
|
xcr1a.1
|
chemokine (C motif) receptor 1a, duplicate 1 |
| chr20_+_52458765 | 0.14 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr9_+_9441453 | 0.14 |
ENSDART00000081859
ENSDART00000188567 ENSDART00000143101 |
maats1
|
MYCBP-associated, testis expressed 1 |
| chr2_+_14992879 | 0.13 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr19_+_4916233 | 0.13 |
ENSDART00000159512
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr2_-_17235891 | 0.13 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr1_-_7603734 | 0.13 |
ENSDART00000009315
|
mxb
|
myxovirus (influenza) resistance B |
| chr1_-_56213723 | 0.13 |
ENSDART00000142505
ENSDART00000137237 |
si:dkey-76b14.2
|
si:dkey-76b14.2 |
| chr22_-_10605045 | 0.13 |
ENSDART00000184812
|
bap1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr1_+_46194333 | 0.13 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr15_-_19677511 | 0.13 |
ENSDART00000043743
|
si:dkey-4p15.3
|
si:dkey-4p15.3 |
| chr2_-_19520324 | 0.13 |
ENSDART00000079877
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr13_+_45431660 | 0.12 |
ENSDART00000099950
|
syf2
|
SYF2 pre-mRNA-splicing factor |
| chr2_-_19520690 | 0.12 |
ENSDART00000133559
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr21_-_8153165 | 0.12 |
ENSDART00000182580
|
CABZ01074363.1
|
|
| chr22_+_2403068 | 0.12 |
ENSDART00000132925
ENSDART00000132569 |
zgc:112977
|
zgc:112977 |
| chr16_+_20934353 | 0.12 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr7_+_18075504 | 0.12 |
ENSDART00000173689
|
si:ch73-40a2.1
|
si:ch73-40a2.1 |
| chr14_+_15597049 | 0.12 |
ENSDART00000159732
|
si:dkey-203a12.8
|
si:dkey-203a12.8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of cdx1a+cdx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.4 | 2.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 0.8 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 0.6 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 0.7 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.2 | 0.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.6 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.7 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 0.9 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.5 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.7 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 0.9 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 2.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.5 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.1 | 0.7 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.9 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.4 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.7 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.4 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 0.2 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.1 | 0.2 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.6 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.2 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.2 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.1 | 1.1 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 1.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.6 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.6 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0003249 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.4 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.3 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.7 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.0 | 0.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 1.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.1 | GO:0010481 | ventriculo bulbo valve development(GO:0003173) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.4 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.3 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.8 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.1 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.7 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.0 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0043596 | nuclear replication fork(GO:0043596) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 0.8 | GO:1990931 | RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.2 | 0.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 0.9 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.2 | 0.5 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 0.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 2.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.5 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.7 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.5 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 0.4 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.9 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.3 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0031834 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 1.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.5 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 1.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 1.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |