Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
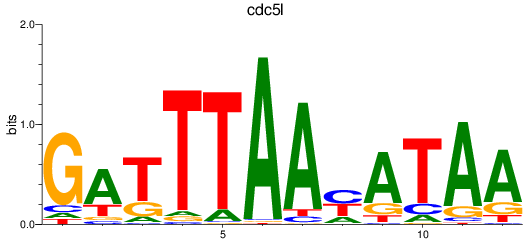
Results for cdc5l
Z-value: 0.61
Transcription factors associated with cdc5l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cdc5l
|
ENSDARG00000043797 | CDC5 cell division cycle 5-like (S. pombe) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cdc5l | dr11_v1_chr17_+_5061135_5061135 | -0.24 | 3.3e-01 | Click! |
Activity profile of cdc5l motif
Sorted Z-values of cdc5l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_42308955 | 1.56 |
ENSDART00000136683
|
pimr201
|
Pim proto-oncogene, serine/threonine kinase, related 201 |
| chr10_+_439692 | 1.12 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr16_-_29557338 | 1.05 |
ENSDART00000058888
|
hormad1
|
HORMA domain containing 1 |
| chr24_-_2959091 | 0.97 |
ENSDART00000170525
ENSDART00000162143 |
cidea
|
cell death-inducing DFFA-like effector a |
| chr22_+_3223489 | 0.96 |
ENSDART00000082011
|
lim2.2
|
lens intrinsic membrane protein 2.2 |
| chr5_-_41494831 | 0.95 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr14_-_33360344 | 0.91 |
ENSDART00000181291
|
pimr117
|
Pim proto-oncogene, serine/threonine kinase, related 117 |
| chr2_-_38000276 | 0.90 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr25_-_19585010 | 0.88 |
ENSDART00000021340
|
sycp3
|
synaptonemal complex protein 3 |
| chr24_+_22022109 | 0.85 |
ENSDART00000133686
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr24_+_13277573 | 0.85 |
ENSDART00000137886
|
si:ch211-171b20.3
|
si:ch211-171b20.3 |
| chr2_-_45691128 | 0.84 |
ENSDART00000125406
|
fam102ba
|
family with sequence similarity 102, member B, a |
| chr14_-_33387288 | 0.82 |
ENSDART00000171388
|
pimr119
|
Pim proto-oncogene, serine/threonine kinase, related 119 |
| chr15_+_1148074 | 0.81 |
ENSDART00000152638
ENSDART00000152466 ENSDART00000188011 |
mlf1
|
myeloid leukemia factor 1 |
| chr21_-_29990574 | 0.78 |
ENSDART00000185800
|
si:ch73-190f9.4
|
si:ch73-190f9.4 |
| chr8_+_17933475 | 0.77 |
ENSDART00000100651
|
erich3
|
glutamate-rich 3 |
| chr1_-_10806625 | 0.77 |
ENSDART00000139749
|
si:ch73-222h13.1
|
si:ch73-222h13.1 |
| chr24_+_22021675 | 0.76 |
ENSDART00000081234
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr22_-_15562933 | 0.75 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr3_+_62126981 | 0.74 |
ENSDART00000060527
|
drc3
|
dynein regulatory complex subunit 3 |
| chr4_+_912563 | 0.73 |
ENSDART00000103631
|
ripply2
|
ripply transcriptional repressor 2 |
| chr19_+_34274504 | 0.72 |
ENSDART00000132046
|
si:ch211-9n13.3
|
si:ch211-9n13.3 |
| chr16_-_35579200 | 0.72 |
ENSDART00000162172
|
scmh1
|
Scm polycomb group protein homolog 1 |
| chr24_+_39443874 | 0.72 |
ENSDART00000142998
|
dnah3
|
dynein axonemal heavy chain 3 |
| chr23_+_27352072 | 0.72 |
ENSDART00000144762
|
si:dkey-9p24.5
|
si:dkey-9p24.5 |
| chr14_-_33407997 | 0.72 |
ENSDART00000163296
|
pimr118
|
Pim proto-oncogene, serine/threonine kinase, related 118 |
| chr5_-_29671586 | 0.69 |
ENSDART00000098336
|
spaca9
|
sperm acrosome associated 9 |
| chr24_-_26310854 | 0.66 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr20_+_35438300 | 0.66 |
ENSDART00000102504
ENSDART00000153249 |
tdrd6
|
tudor domain containing 6 |
| chr25_-_19584735 | 0.66 |
ENSDART00000137930
|
sycp3
|
synaptonemal complex protein 3 |
| chr21_-_29477624 | 0.66 |
ENSDART00000180110
|
BX537120.1
|
|
| chr21_-_29940453 | 0.65 |
ENSDART00000182862
|
CU104716.1
|
|
| chr21_-_41870029 | 0.65 |
ENSDART00000182035
|
endou2
|
endonuclease, polyU-specific 2 |
| chr6_+_12968101 | 0.64 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr21_-_29735366 | 0.63 |
ENSDART00000181668
|
CR847571.2
|
|
| chr21_-_29689141 | 0.61 |
ENSDART00000100876
|
CR847571.1
|
|
| chr4_-_13567387 | 0.60 |
ENSDART00000132971
ENSDART00000102010 |
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr17_-_52521002 | 0.60 |
ENSDART00000114931
|
prox2
|
prospero homeobox 2 |
| chr3_+_62161184 | 0.60 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr19_+_636886 | 0.59 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr25_-_4192637 | 0.59 |
ENSDART00000153832
|
ppp1r32
|
protein phosphatase 1, regulatory subunit 32 |
| chr5_+_29671681 | 0.58 |
ENSDART00000043096
|
ak8
|
adenylate kinase 8 |
| chr16_+_4838808 | 0.58 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr25_+_5035343 | 0.57 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr14_-_33396934 | 0.57 |
ENSDART00000158606
|
pimr111
|
Pim proto-oncogene, serine/threonine kinase, related 111 |
| chr25_-_4482449 | 0.57 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr24_+_24064562 | 0.55 |
ENSDART00000144394
|
zgc:112408
|
zgc:112408 |
| chr23_+_45512825 | 0.55 |
ENSDART00000064846
|
prelid1b
|
PRELI domain containing 1b |
| chr4_-_56068511 | 0.55 |
ENSDART00000168345
|
znf1133
|
zinc finger protein 1133 |
| chr24_+_12945803 | 0.55 |
ENSDART00000005105
|
psme1
|
proteasome activator subunit 1 |
| chr21_+_13577227 | 0.55 |
ENSDART00000146283
|
si:dkey-248f6.3
|
si:dkey-248f6.3 |
| chr8_-_11546175 | 0.55 |
ENSDART00000081909
|
si:ch211-248e11.2
|
si:ch211-248e11.2 |
| chr2_+_22531185 | 0.54 |
ENSDART00000171959
|
hfm1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr1_+_57269441 | 0.54 |
ENSDART00000149688
|
mycbpap
|
mycbp associated protein |
| chr1_+_50538839 | 0.54 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr21_+_20771082 | 0.53 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr21_-_29533723 | 0.52 |
ENSDART00000193554
|
BX537120.2
|
|
| chr25_-_15245475 | 0.51 |
ENSDART00000142943
|
ccdc73
|
coiled-coil domain containing 73 |
| chr17_+_27545183 | 0.51 |
ENSDART00000129392
|
pacrg
|
PARK2 co-regulated |
| chr7_-_59311165 | 0.50 |
ENSDART00000171105
|
m1ap
|
meiosis 1 associated protein |
| chr13_+_36764715 | 0.50 |
ENSDART00000111832
ENSDART00000085230 |
atl1
|
atlastin GTPase 1 |
| chr8_-_4431971 | 0.49 |
ENSDART00000141728
ENSDART00000133519 |
rad9b
|
RAD9 checkpoint clamp component B |
| chr17_+_25414033 | 0.49 |
ENSDART00000001691
|
tdh2
|
L-threonine dehydrogenase 2 |
| chr19_-_3127271 | 0.49 |
ENSDART00000146928
ENSDART00000134197 |
si:ch211-80h18.1
|
si:ch211-80h18.1 |
| chr8_-_29719393 | 0.49 |
ENSDART00000077635
|
si:ch211-103n10.5
|
si:ch211-103n10.5 |
| chr18_+_1145571 | 0.48 |
ENSDART00000135055
|
rec114
|
REC114 meiotic recombination protein |
| chr3_-_2593859 | 0.48 |
ENSDART00000143826
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr12_+_20641102 | 0.47 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr17_-_36936856 | 0.46 |
ENSDART00000010274
ENSDART00000188887 |
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr23_-_3409140 | 0.46 |
ENSDART00000002309
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr24_+_26345427 | 0.46 |
ENSDART00000089756
|
lrrc34
|
leucine rich repeat containing 34 |
| chr14_-_33370378 | 0.46 |
ENSDART00000180068
|
pimr120
|
Pim proto-oncogene, serine/threonine kinase, related 120 |
| chr25_-_7764083 | 0.46 |
ENSDART00000179800
ENSDART00000181858 |
phf21ab
|
PHD finger protein 21Ab |
| chr5_+_53824959 | 0.45 |
ENSDART00000169565
|
sat2b
|
spermidine/spermine N1-acetyltransferase family member 2b |
| chr14_+_6991142 | 0.45 |
ENSDART00000157635
ENSDART00000059918 |
hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr4_+_14900042 | 0.45 |
ENSDART00000018261
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr24_-_26399623 | 0.45 |
ENSDART00000112317
|
zgc:194621
|
zgc:194621 |
| chr3_-_16537441 | 0.45 |
ENSDART00000080749
ENSDART00000133824 |
eps8l1
|
eps8-like1 |
| chr14_-_28432699 | 0.44 |
ENSDART00000054062
|
nek12
|
NIMA-related kinase 12 |
| chr16_-_25085327 | 0.43 |
ENSDART00000077661
|
prss1
|
protease, serine 1 |
| chr25_-_9013963 | 0.43 |
ENSDART00000073912
|
rag2
|
recombination activating gene 2 |
| chr17_-_31695217 | 0.43 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr1_+_2129164 | 0.43 |
ENSDART00000074923
ENSDART00000124534 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr19_-_19339285 | 0.43 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr4_-_11580948 | 0.43 |
ENSDART00000049066
|
net1
|
neuroepithelial cell transforming 1 |
| chr22_-_20309283 | 0.42 |
ENSDART00000182125
ENSDART00000048775 |
si:dkey-110c1.10
|
si:dkey-110c1.10 |
| chr11_+_1533097 | 0.42 |
ENSDART00000066191
|
ift52
|
intraflagellar transport 52 homolog (Chlamydomonas) |
| chr16_-_12809873 | 0.42 |
ENSDART00000146997
ENSDART00000178291 ENSDART00000007842 |
isoc2
|
isochorismatase domain containing 2 |
| chr21_-_32301109 | 0.42 |
ENSDART00000139890
|
clk4b
|
CDC-like kinase 4b |
| chr4_-_17642168 | 0.41 |
ENSDART00000007030
|
klhl42
|
kelch-like family, member 42 |
| chr2_+_6181383 | 0.41 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr16_-_1757521 | 0.41 |
ENSDART00000124660
|
ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr19_-_20163197 | 0.40 |
ENSDART00000148179
ENSDART00000151646 |
fam221a
|
family with sequence similarity 221, member A |
| chr10_+_6625785 | 0.40 |
ENSDART00000148433
|
si:ch211-57m13.3
|
si:ch211-57m13.3 |
| chr19_+_25497541 | 0.40 |
ENSDART00000144300
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr18_+_21122818 | 0.40 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr24_-_3426620 | 0.40 |
ENSDART00000184346
|
nck1b
|
NCK adaptor protein 1b |
| chr20_-_1174266 | 0.39 |
ENSDART00000023304
|
gabrr2b
|
gamma-aminobutyric acid (GABA) A receptor, rho 2b |
| chr18_+_36914221 | 0.39 |
ENSDART00000147656
|
si:ch211-238g23.1
|
si:ch211-238g23.1 |
| chr1_-_44701313 | 0.38 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr10_-_44026369 | 0.38 |
ENSDART00000185456
|
crybb1
|
crystallin, beta B1 |
| chr21_+_27382893 | 0.38 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr17_-_26926577 | 0.38 |
ENSDART00000050202
|
rcan3
|
regulator of calcineurin 3 |
| chr7_-_52388090 | 0.38 |
ENSDART00000174104
|
wdr93
|
WD repeat domain 93 |
| chr23_-_19558556 | 0.38 |
ENSDART00000134012
|
dnah12
|
dynein, axonemal, heavy chain 12 |
| chr14_-_41285392 | 0.38 |
ENSDART00000147389
|
tmem35
|
transmembrane protein 35 |
| chr23_+_32011768 | 0.37 |
ENSDART00000053509
|
plagx
|
pleiomorphic adenoma gene X |
| chr6_+_46259950 | 0.37 |
ENSDART00000032326
|
zgc:162324
|
zgc:162324 |
| chr10_-_5844915 | 0.36 |
ENSDART00000185929
|
ankrd55
|
ankyrin repeat domain 55 |
| chr1_+_12009673 | 0.36 |
ENSDART00000080100
|
slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr8_-_12403077 | 0.36 |
ENSDART00000142150
|
phf19
|
PHD finger protein 19 |
| chr1_-_51734524 | 0.35 |
ENSDART00000109640
ENSDART00000122628 |
junba
|
JunB proto-oncogene, AP-1 transcription factor subunit a |
| chr10_+_31222433 | 0.35 |
ENSDART00000185080
|
tmem218
|
transmembrane protein 218 |
| chr22_-_10580194 | 0.35 |
ENSDART00000105848
|
SHISA4 (1 of many)
|
si:dkey-42i9.7 |
| chr24_+_40860320 | 0.35 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr22_-_31059670 | 0.34 |
ENSDART00000022445
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr7_-_16598212 | 0.34 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr13_+_4409294 | 0.34 |
ENSDART00000146437
|
si:ch211-130h14.4
|
si:ch211-130h14.4 |
| chr7_+_20467549 | 0.34 |
ENSDART00000173724
|
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr4_-_71110826 | 0.34 |
ENSDART00000167431
|
si:dkeyp-80d11.1
|
si:dkeyp-80d11.1 |
| chr5_-_69425224 | 0.33 |
ENSDART00000158096
|
FQ311924.1
|
|
| chr6_-_49063085 | 0.33 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr20_-_29787192 | 0.33 |
ENSDART00000125348
ENSDART00000048759 |
id2b
|
inhibitor of DNA binding 2b |
| chr24_-_38657683 | 0.32 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr22_+_1911269 | 0.32 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr7_-_24994722 | 0.32 |
ENSDART00000131671
|
rcor2
|
REST corepressor 2 |
| chr24_+_26345609 | 0.32 |
ENSDART00000186844
|
lrrc34
|
leucine rich repeat containing 34 |
| chr17_+_6563307 | 0.32 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr13_-_6218248 | 0.32 |
ENSDART00000159052
|
si:zfos-1056e6.1
|
si:zfos-1056e6.1 |
| chr8_-_6918290 | 0.32 |
ENSDART00000138259
ENSDART00000142496 |
asb6
|
ankyrin repeat and SOCS box containing 6 |
| chr11_+_16152316 | 0.32 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr1_+_25178106 | 0.32 |
ENSDART00000054265
ENSDART00000141648 |
mnd1
|
meiotic nuclear divisions 1 homolog (S. cerevisiae) |
| chr19_+_5604241 | 0.31 |
ENSDART00000011025
|
wipf2b
|
WAS/WASL interacting protein family, member 2b |
| chr21_+_18925318 | 0.31 |
ENSDART00000136182
|
si:ch211-222n4.2
|
si:ch211-222n4.2 |
| chr20_+_263056 | 0.31 |
ENSDART00000132669
|
tube1
|
tubulin, epsilon 1 |
| chr24_+_12989727 | 0.31 |
ENSDART00000126842
ENSDART00000129309 |
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr2_-_42958619 | 0.31 |
ENSDART00000144317
|
oc90
|
otoconin 90 |
| chr22_+_15898221 | 0.31 |
ENSDART00000062587
|
klf2a
|
Kruppel-like factor 2a |
| chr18_+_20838786 | 0.31 |
ENSDART00000138692
|
ttc23
|
tetratricopeptide repeat domain 23 |
| chr3_+_28831450 | 0.31 |
ENSDART00000055422
|
flr
|
fleer |
| chr4_+_35553514 | 0.31 |
ENSDART00000182938
|
CR847906.1
|
|
| chr1_+_49651016 | 0.31 |
ENSDART00000074380
ENSDART00000101017 |
tsga10
|
testis specific, 10 |
| chr24_+_23742690 | 0.31 |
ENSDART00000130162
|
TCF24
|
transcription factor 24 |
| chr15_-_20024205 | 0.31 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr1_-_36151377 | 0.30 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr7_-_51757085 | 0.30 |
ENSDART00000167747
|
leap2
|
liver-expressed antimicrobial peptide 2 |
| chr7_-_22956889 | 0.30 |
ENSDART00000101447
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr23_+_37086159 | 0.30 |
ENSDART00000074407
|
cptp
|
ceramide-1-phosphate transfer protein |
| chr16_-_21540077 | 0.30 |
ENSDART00000078790
|
stk31
|
serine/threonine kinase 31 |
| chr24_-_20369604 | 0.30 |
ENSDART00000143174
|
dlec1
|
deleted in lung and esophageal cancer 1 |
| chr1_-_7570181 | 0.30 |
ENSDART00000103588
|
mxa
|
myxovirus (influenza) resistance A |
| chr12_+_48220584 | 0.29 |
ENSDART00000164392
|
lrrc20
|
leucine rich repeat containing 20 |
| chr6_-_49526510 | 0.29 |
ENSDART00000128025
|
rps26l
|
ribosomal protein S26, like |
| chr6_+_9893554 | 0.29 |
ENSDART00000064979
|
pimr74
|
Pim proto-oncogene, serine/threonine kinase, related 74 |
| chr15_+_19293744 | 0.29 |
ENSDART00000184994
ENSDART00000123815 |
jam3a
|
junctional adhesion molecule 3a |
| chr11_-_16395956 | 0.29 |
ENSDART00000115085
|
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr14_+_15622817 | 0.29 |
ENSDART00000158624
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr2_-_42960353 | 0.29 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr5_+_70155935 | 0.29 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr4_+_686157 | 0.29 |
ENSDART00000169748
|
fbxo7
|
F-box protein 7 |
| chr25_+_31267268 | 0.28 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr16_-_25699554 | 0.28 |
ENSDART00000155899
ENSDART00000114495 |
ddx61
|
DEAD (Asp-Glu-Ala-Asp) box helicase 61 |
| chr7_+_25053331 | 0.28 |
ENSDART00000173998
|
si:dkey-23i12.7
|
si:dkey-23i12.7 |
| chr13_-_37620091 | 0.28 |
ENSDART00000135875
ENSDART00000193270 ENSDART00000018064 |
zgc:152791
|
zgc:152791 |
| chr5_-_26834511 | 0.28 |
ENSDART00000136713
ENSDART00000192932 ENSDART00000113246 |
si:ch211-102c2.4
|
si:ch211-102c2.4 |
| chr4_+_3358383 | 0.27 |
ENSDART00000075320
|
nampta
|
nicotinamide phosphoribosyltransferase a |
| chr22_-_15593824 | 0.27 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr1_+_34224360 | 0.27 |
ENSDART00000192938
|
arl6
|
ADP-ribosylation factor-like 6 |
| chr12_+_16452575 | 0.27 |
ENSDART00000058665
|
kif20bb
|
kinesin family member 20Bb |
| chr7_+_31838320 | 0.27 |
ENSDART00000144679
ENSDART00000174217 ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr23_-_25798099 | 0.27 |
ENSDART00000041833
|
fitm2
|
fat storage-inducing transmembrane protein 2 |
| chr15_+_15390882 | 0.27 |
ENSDART00000062024
|
ca4b
|
carbonic anhydrase IV b |
| chr15_-_5815006 | 0.26 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr17_-_24439672 | 0.26 |
ENSDART00000155020
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr1_+_2190714 | 0.26 |
ENSDART00000132126
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr3_-_42125655 | 0.26 |
ENSDART00000040753
|
nudt1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
| chr6_+_55174744 | 0.26 |
ENSDART00000023562
|
SYT2
|
synaptotagmin 2 |
| chr1_-_7894255 | 0.26 |
ENSDART00000167126
ENSDART00000145460 |
radil
|
Ras association and DIL domains |
| chr12_+_16284086 | 0.25 |
ENSDART00000013360
ENSDART00000141169 |
ppp1r3cb
|
protein phosphatase 1, regulatory subunit 3Cb |
| chr15_+_28096152 | 0.25 |
ENSDART00000100293
ENSDART00000140092 |
crybb1l3
|
crystallin, beta B1, like 3 |
| chr14_+_8475007 | 0.25 |
ENSDART00000148210
|
dnajc4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr10_+_24627683 | 0.25 |
ENSDART00000112652
|
slc46a3
|
solute carrier family 46, member 3 |
| chr23_+_44497701 | 0.25 |
ENSDART00000149903
|
si:ch73-375g18.1
|
si:ch73-375g18.1 |
| chr21_+_10076203 | 0.25 |
ENSDART00000190383
|
CABZ01043506.1
|
|
| chr21_-_5393125 | 0.25 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr16_+_23398369 | 0.25 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr4_-_779796 | 0.25 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr24_-_22756508 | 0.25 |
ENSDART00000035409
ENSDART00000146247 |
zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr5_+_45918680 | 0.25 |
ENSDART00000036242
|
ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr16_+_8668519 | 0.24 |
ENSDART00000172893
|
ccr2
|
chemokine (C-C motif) receptor 2 |
| chr17_-_14700889 | 0.24 |
ENSDART00000179975
|
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr8_+_45003659 | 0.24 |
ENSDART00000132663
|
si:ch211-163b2.4
|
si:ch211-163b2.4 |
| chr25_-_15040369 | 0.24 |
ENSDART00000159342
ENSDART00000166490 |
pax6a
|
paired box 6a |
| chr3_-_40202607 | 0.24 |
ENSDART00000074757
|
znf598
|
zinc finger protein 598 |
| chr11_-_8269227 | 0.24 |
ENSDART00000127202
ENSDART00000158079 ENSDART00000158748 |
si:cabz01021067.1
pimr202
|
si:cabz01021067.1 Pim proto-oncogene, serine/threonine kinase, related 202 |
| chr3_-_45308394 | 0.24 |
ENSDART00000155324
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr4_+_5205265 | 0.24 |
ENSDART00000067382
|
galnt8b.1
|
polypeptide N-acetylgalactosaminyltransferase 8b, tandem duplicate 1 |
| chr6_-_12456077 | 0.24 |
ENSDART00000190107
|
gpd2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr2_+_28453338 | 0.24 |
ENSDART00000020456
|
mmp15b
|
matrix metallopeptidase 15b |
| chr10_+_26634472 | 0.24 |
ENSDART00000143655
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
Network of associatons between targets according to the STRING database.
First level regulatory network of cdc5l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.2 | 0.6 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 0.5 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.2 | 1.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.4 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 0.6 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.5 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 0.5 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 1.1 | GO:0030719 | P granule organization(GO:0030719) |
| 0.1 | 0.3 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.1 | 0.5 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.6 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.3 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.1 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 1.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.3 | GO:1902883 | negative regulation of cellular response to oxidative stress(GO:1900408) negative regulation of response to oxidative stress(GO:1902883) regulation of oxidative stress-induced cell death(GO:1903201) negative regulation of oxidative stress-induced cell death(GO:1903202) |
| 0.1 | 0.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.1 | 0.5 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.5 | GO:0036372 | opsin transport(GO:0036372) |
| 0.1 | 0.2 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.1 | 0.6 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.3 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.5 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.3 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 0.5 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.4 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 0.2 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.7 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.5 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.4 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.0 | 0.3 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.3 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.4 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.1 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.1 | GO:0044803 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.5 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 1.0 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:1902267 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.6 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.7 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.5 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.1 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 0.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.2 | GO:1903426 | regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.7 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.8 | GO:0033500 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0021519 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.0 | 0.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 7.3 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.6 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.5 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.9 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.2 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.4 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.0 | 0.1 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.2 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.0 | 0.1 | GO:1901825 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.0 | 0.1 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) positive regulation of lipid transport(GO:0032370) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.4 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.0 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.2 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.1 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.0 | GO:1904353 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 0.6 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 1.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.4 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.1 | 0.5 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.3 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.1 | 0.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.2 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.1 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.0 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.4 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.2 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:0030689 | Noc complex(GO:0030689) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.2 | 0.5 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 0.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.6 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.6 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.5 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.5 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 1.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.5 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.1 | 0.3 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.2 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.1 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.0 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.3 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.5 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 1.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.6 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 1.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |