Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
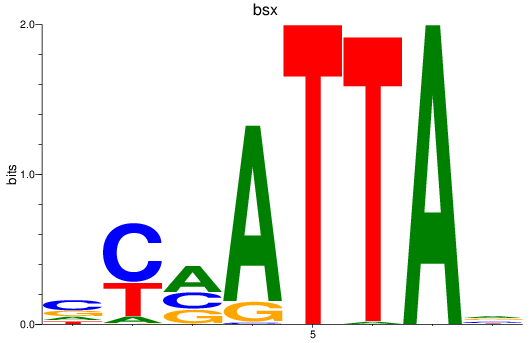
Results for bsx
Z-value: 0.63
Transcription factors associated with bsx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bsx
|
ENSDARG00000068976 | brain-specific homeobox |
|
bsx
|
ENSDARG00000110782 | brain-specific homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bsx | dr11_v1_chr10_-_29892486_29892486 | -0.27 | 2.8e-01 | Click! |
Activity profile of bsx motif
Sorted Z-values of bsx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_18130300 | 2.47 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr20_-_23426339 | 1.85 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr11_+_18183220 | 1.74 |
ENSDART00000113468
|
LO018315.10
|
|
| chr11_+_18157260 | 1.69 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr11_+_18175893 | 1.62 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr12_-_42368296 | 1.42 |
ENSDART00000171075
|
zgc:111868
|
zgc:111868 |
| chr3_-_53091946 | 1.33 |
ENSDART00000187297
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr24_-_10014512 | 1.10 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr3_-_53092509 | 1.06 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr3_-_3448095 | 1.01 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr21_-_43666420 | 1.00 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr11_-_6452444 | 0.99 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr24_-_10021341 | 0.96 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr16_-_24661526 | 0.95 |
ENSDART00000058959
|
pon3.2
|
paraoxonase 3, tandem duplicate 2 |
| chr5_-_9216758 | 0.95 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr24_+_34069675 | 0.88 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr10_+_35554219 | 0.79 |
ENSDART00000077373
|
zdhhc20a
|
zinc finger, DHHC-type containing 20a |
| chr3_+_28860283 | 0.77 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr1_-_513762 | 0.74 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr8_+_10304981 | 0.72 |
ENSDART00000160766
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr2_-_38287987 | 0.70 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr24_+_40860320 | 0.69 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr10_-_44560165 | 0.67 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr16_-_17197546 | 0.65 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr6_+_13806466 | 0.63 |
ENSDART00000043522
|
tmem198b
|
transmembrane protein 198b |
| chr24_-_37640705 | 0.63 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr24_+_26134209 | 0.60 |
ENSDART00000038824
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr13_+_21768447 | 0.55 |
ENSDART00000100941
|
chchd1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chr13_-_36911118 | 0.53 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr20_-_40758410 | 0.53 |
ENSDART00000183031
|
cx34.5
|
connexin 34.5 |
| chr2_+_8780443 | 0.53 |
ENSDART00000137768
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr12_-_32073611 | 0.52 |
ENSDART00000153369
ENSDART00000086531 |
cox11
|
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr14_-_28567845 | 0.51 |
ENSDART00000126095
|
insb
|
preproinsulin b |
| chr3_+_56366395 | 0.50 |
ENSDART00000154367
|
cacng5b
|
calcium channel, voltage-dependent, gamma subunit 5b |
| chr13_+_17468161 | 0.49 |
ENSDART00000008906
|
znf503
|
zinc finger protein 503 |
| chr17_+_24821627 | 0.47 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr25_-_27621268 | 0.46 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr1_-_55248496 | 0.45 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr10_-_21362071 | 0.45 |
ENSDART00000125167
|
avd
|
avidin |
| chr21_-_2209714 | 0.45 |
ENSDART00000163659
|
zgc:162971
|
zgc:162971 |
| chr15_+_41027466 | 0.44 |
ENSDART00000075940
|
mtnr1ba
|
melatonin receptor type 1Ba |
| chr19_+_7152966 | 0.44 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr19_+_15521997 | 0.43 |
ENSDART00000003164
|
ppp1r8a
|
protein phosphatase 1, regulatory subunit 8a |
| chr3_-_26244256 | 0.43 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr1_-_45662774 | 0.43 |
ENSDART00000042158
|
serhl
|
serine hydrolase-like |
| chr10_-_21362320 | 0.43 |
ENSDART00000189789
|
avd
|
avidin |
| chr10_-_25217347 | 0.42 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr21_-_36396334 | 0.42 |
ENSDART00000183627
|
mrpl22
|
mitochondrial ribosomal protein L22 |
| chr6_-_35046735 | 0.41 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr9_-_24031461 | 0.41 |
ENSDART00000021218
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr6_+_58832155 | 0.40 |
ENSDART00000144842
|
dctn2
|
dynactin 2 (p50) |
| chr13_-_3370638 | 0.40 |
ENSDART00000029649
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr16_+_7242610 | 0.37 |
ENSDART00000081477
|
sri
|
sorcin |
| chr10_-_23099809 | 0.37 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr2_+_38892631 | 0.37 |
ENSDART00000134040
|
si:ch211-119o8.4
|
si:ch211-119o8.4 |
| chr17_+_16046132 | 0.37 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr13_-_9841806 | 0.36 |
ENSDART00000101949
|
sfxn4
|
sideroflexin 4 |
| chr6_+_52212927 | 0.36 |
ENSDART00000143458
|
ywhaba
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide a |
| chr11_-_16152400 | 0.35 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr1_-_45049603 | 0.35 |
ENSDART00000023336
|
rps6
|
ribosomal protein S6 |
| chr7_+_17106160 | 0.35 |
ENSDART00000190048
ENSDART00000180004 ENSDART00000013409 |
prmt3
|
protein arginine methyltransferase 3 |
| chr1_-_12064715 | 0.35 |
ENSDART00000143628
ENSDART00000103406 |
pla2g12a
|
phospholipase A2, group XIIA |
| chr9_-_24030795 | 0.35 |
ENSDART00000144163
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr8_+_36500061 | 0.35 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr11_-_36350876 | 0.34 |
ENSDART00000146495
ENSDART00000020655 |
psma5
|
proteasome subunit alpha 5 |
| chr6_-_46474483 | 0.33 |
ENSDART00000155761
|
rdh20
|
retinol dehydrogenase 20 |
| chr14_+_24935131 | 0.33 |
ENSDART00000170871
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr20_-_28433990 | 0.33 |
ENSDART00000182824
ENSDART00000193381 |
wdr21
|
WD repeat domain 21 |
| chr4_+_14981854 | 0.33 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr16_+_33902006 | 0.33 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr24_-_36271352 | 0.33 |
ENSDART00000153682
ENSDART00000155892 |
si:ch211-40k21.5
|
si:ch211-40k21.5 |
| chr19_-_5103141 | 0.33 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr18_-_29896367 | 0.32 |
ENSDART00000191303
|
cmc2
|
C-x(9)-C motif containing 2 |
| chr12_-_48477031 | 0.32 |
ENSDART00000105176
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr24_-_26945390 | 0.32 |
ENSDART00000123354
|
msl2b
|
male-specific lethal 2 homolog b (Drosophila) |
| chr17_+_16046314 | 0.32 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr20_+_18260358 | 0.32 |
ENSDART00000187734
ENSDART00000191947 ENSDART00000057039 |
taf4b
|
TAF4B RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr21_+_42226113 | 0.30 |
ENSDART00000170362
|
GABRB2 (1 of many)
|
gamma-aminobutyric acid type A receptor beta2 subunit |
| chr2_+_10007113 | 0.30 |
ENSDART00000155213
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr25_-_32869794 | 0.29 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr23_-_20051369 | 0.29 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr23_+_39695827 | 0.29 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr17_+_37253706 | 0.29 |
ENSDART00000076004
|
tmem62
|
transmembrane protein 62 |
| chr16_-_15988320 | 0.29 |
ENSDART00000160883
|
CABZ01060453.1
|
|
| chr20_-_45060241 | 0.29 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr19_+_12406583 | 0.28 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr12_+_22580579 | 0.28 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr3_-_32596394 | 0.28 |
ENSDART00000103239
|
tspan4b
|
tetraspanin 4b |
| chr13_-_11984867 | 0.28 |
ENSDART00000157538
|
npm3
|
nucleophosmin/nucleoplasmin, 3 |
| chr5_-_19006290 | 0.27 |
ENSDART00000137022
|
golga3
|
golgin A3 |
| chr25_-_36370292 | 0.27 |
ENSDART00000152766
|
CR354435.1
|
Histone H2B 1/2 |
| chr12_+_20352400 | 0.27 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr7_+_24528430 | 0.27 |
ENSDART00000133022
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr5_+_1079423 | 0.27 |
ENSDART00000172231
|
si:zfos-128g4.2
|
si:zfos-128g4.2 |
| chr21_+_25777425 | 0.27 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr20_+_2134816 | 0.27 |
ENSDART00000039249
|
l3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr6_+_29707129 | 0.26 |
ENSDART00000023549
ENSDART00000164849 |
phb2b
|
prohibitin 2b |
| chr25_+_32755485 | 0.26 |
ENSDART00000162188
|
etfa
|
electron-transfer-flavoprotein, alpha polypeptide |
| chr3_-_41791178 | 0.25 |
ENSDART00000049687
|
grifin
|
galectin-related inter-fiber protein |
| chr10_+_25947946 | 0.25 |
ENSDART00000064393
|
ufm1
|
ubiquitin-fold modifier 1 |
| chr3_+_17537352 | 0.25 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr15_-_16177603 | 0.25 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr10_+_23099890 | 0.25 |
ENSDART00000135890
|
LTN1
|
si:dkey-175g6.5 |
| chr12_+_23912074 | 0.25 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr21_+_37442458 | 0.25 |
ENSDART00000180161
ENSDART00000186950 |
cox7b
|
cytochrome c oxidase subunit VIIb |
| chr3_-_16719244 | 0.25 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr20_-_37813863 | 0.25 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr5_+_68807170 | 0.25 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr8_-_53166975 | 0.25 |
ENSDART00000114683
|
rbsn
|
rabenosyn, RAB effector |
| chr4_-_14191434 | 0.24 |
ENSDART00000142374
ENSDART00000136730 |
pus7l
|
pseudouridylate synthase 7-like |
| chr21_+_38732945 | 0.24 |
ENSDART00000076157
|
rab24
|
RAB24, member RAS oncogene family |
| chr7_-_54679595 | 0.24 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr5_-_68333081 | 0.24 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr10_-_34002185 | 0.24 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr18_-_26894732 | 0.23 |
ENSDART00000147735
ENSDART00000188938 |
BX470164.1
si:dkey-24l11.2
|
si:dkey-24l11.2 |
| chr19_-_10330778 | 0.23 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr13_+_2625150 | 0.23 |
ENSDART00000164177
|
plpp4
|
phospholipid phosphatase 4 |
| chr25_-_23583101 | 0.23 |
ENSDART00000149107
ENSDART00000103704 ENSDART00000184903 |
nap1l4a
|
nucleosome assembly protein 1-like 4a |
| chr19_-_5103313 | 0.22 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr11_-_1550709 | 0.22 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr7_-_23894879 | 0.22 |
ENSDART00000123203
|
vbp1
|
von Hippel-Lindau binding protein 1 |
| chr23_-_35064785 | 0.21 |
ENSDART00000172240
|
BX294434.1
|
|
| chr14_-_24110251 | 0.21 |
ENSDART00000079226
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr11_+_33312601 | 0.21 |
ENSDART00000188024
|
cntnap5l
|
contactin associated protein-like 5 like |
| chr3_+_6469754 | 0.21 |
ENSDART00000185809
|
NUP85 (1 of many)
|
nucleoporin 85 |
| chr4_-_2380173 | 0.21 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr1_+_11977426 | 0.21 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr3_+_32365811 | 0.20 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr12_+_10116912 | 0.20 |
ENSDART00000189630
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr10_+_518546 | 0.20 |
ENSDART00000128275
|
npffr1l3
|
neuropeptide FF receptor 1 like 3 |
| chr25_-_29087925 | 0.20 |
ENSDART00000171758
|
rpp25a
|
ribonuclease P and MRP subunit p25, a |
| chr8_-_20243389 | 0.20 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr9_+_50600355 | 0.20 |
ENSDART00000187567
|
fign
|
fidgetin |
| chr16_+_42471455 | 0.20 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr21_-_2322102 | 0.20 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr24_-_24111674 | 0.19 |
ENSDART00000024606
|
zgc:112982
|
zgc:112982 |
| chr14_+_34490445 | 0.19 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr3_+_18398876 | 0.19 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr19_+_31585917 | 0.18 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr4_-_16833518 | 0.18 |
ENSDART00000179867
|
ldhba
|
lactate dehydrogenase Ba |
| chr23_-_25135046 | 0.18 |
ENSDART00000184844
ENSDART00000103989 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr19_+_5543072 | 0.18 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr5_+_50879545 | 0.18 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr16_+_29509133 | 0.18 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr25_+_35375848 | 0.18 |
ENSDART00000155721
|
ano3
|
anoctamin 3 |
| chr13_+_36984794 | 0.18 |
ENSDART00000137328
|
frmd6
|
FERM domain containing 6 |
| chr1_-_33645967 | 0.18 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr14_+_5936996 | 0.18 |
ENSDART00000097144
ENSDART00000126777 |
kctd8
|
potassium channel tetramerization domain containing 8 |
| chr2_+_26656283 | 0.17 |
ENSDART00000133202
ENSDART00000099208 |
asph
|
aspartate beta-hydroxylase |
| chr10_-_32494499 | 0.17 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr16_+_47207691 | 0.17 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr21_+_34088110 | 0.17 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr22_-_20166660 | 0.17 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr12_+_10115964 | 0.16 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr16_-_32304764 | 0.16 |
ENSDART00000143859
ENSDART00000134381 |
mms22l
|
MMS22-like, DNA repair protein |
| chr21_-_11654422 | 0.16 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr10_-_32494304 | 0.16 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr19_-_205104 | 0.16 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr8_-_37249813 | 0.16 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr7_-_17816175 | 0.16 |
ENSDART00000091272
ENSDART00000173757 |
ecsit
|
ECSIT signalling integrator |
| chr5_+_29794058 | 0.16 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr21_+_17768174 | 0.16 |
ENSDART00000141380
|
rxraa
|
retinoid X receptor, alpha a |
| chr21_+_4313039 | 0.16 |
ENSDART00000141146
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr22_+_9003090 | 0.15 |
ENSDART00000106414
|
rnh1
|
ribonuclease/angiogenin inhibitor 1 |
| chr23_-_35790235 | 0.15 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr1_-_39983730 | 0.15 |
ENSDART00000160066
|
ing2
|
inhibitor of growth family, member 2 |
| chr6_+_47846366 | 0.15 |
ENSDART00000064842
|
padi2
|
peptidyl arginine deiminase, type II |
| chr2_+_37227011 | 0.15 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr6_-_25163722 | 0.15 |
ENSDART00000192225
|
znf326
|
zinc finger protein 326 |
| chr24_-_30843250 | 0.15 |
ENSDART00000162920
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr19_+_31873308 | 0.15 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr11_-_28050559 | 0.15 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr2_+_50608099 | 0.15 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr5_-_31856681 | 0.14 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr25_+_25516016 | 0.14 |
ENSDART00000188980
|
phrf1
|
PHD and ring finger domains 1 |
| chr14_-_33334065 | 0.14 |
ENSDART00000052761
|
rpl39
|
ribosomal protein L39 |
| chr21_-_35325466 | 0.14 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr6_+_28208973 | 0.14 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr11_-_36341028 | 0.14 |
ENSDART00000146093
|
sort1a
|
sortilin 1a |
| chr21_-_11575626 | 0.14 |
ENSDART00000133171
|
cast
|
calpastatin |
| chr17_-_6514962 | 0.14 |
ENSDART00000163514
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
| chr24_+_32668675 | 0.14 |
ENSDART00000156638
ENSDART00000155973 |
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr23_-_4966821 | 0.13 |
ENSDART00000147122
ENSDART00000145760 |
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr11_+_26604224 | 0.13 |
ENSDART00000030453
ENSDART00000168895 ENSDART00000159505 |
dynlrb1
|
dynein, light chain, roadblock-type 1 |
| chr8_+_26253252 | 0.13 |
ENSDART00000142031
|
slc26a6
|
solute carrier family 26, member 6 |
| chr9_-_35875051 | 0.13 |
ENSDART00000013432
|
dnajc28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr21_+_45502621 | 0.13 |
ENSDART00000166719
|
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr2_-_32666174 | 0.13 |
ENSDART00000133660
|
puf60a
|
poly-U binding splicing factor a |
| chr8_+_31435452 | 0.12 |
ENSDART00000145282
|
selenop
|
selenoprotein P |
| chr16_+_39159752 | 0.12 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr5_+_6954162 | 0.12 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr10_+_33744098 | 0.12 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr25_+_25508495 | 0.12 |
ENSDART00000150719
|
phrf1
|
PHD and ring finger domains 1 |
| chr22_-_10586191 | 0.12 |
ENSDART00000148418
|
si:dkey-42i9.16
|
si:dkey-42i9.16 |
| chr15_+_436776 | 0.12 |
ENSDART00000008504
|
med17
|
mediator complex subunit 17 |
| chr16_-_49646625 | 0.12 |
ENSDART00000101629
|
efhb
|
EF-hand domain family, member B |
| chr23_-_19715557 | 0.12 |
ENSDART00000143764
|
rpl10
|
ribosomal protein L10 |
| chr15_-_16704417 | 0.12 |
ENSDART00000155163
|
caln1
|
calneuron 1 |
| chr25_+_18965430 | 0.11 |
ENSDART00000169742
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr19_+_2279051 | 0.11 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of bsx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.3 | 0.8 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.1 | 0.7 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.6 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.4 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.3 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.2 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.3 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.2 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 3.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.3 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.7 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.6 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.3 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.3 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 1.5 | GO:0018198 | peptidyl-cysteine modification(GO:0018198) |
| 0.0 | 0.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.2 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.1 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.0 | 0.5 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.7 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.5 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.2 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 0.3 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.4 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.1 | GO:0051645 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.0 | 2.5 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.3 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.5 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.4 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0060254 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.0 | 0.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.2 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 0.2 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.0 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.0 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.3 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 3.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.2 | 0.7 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.2 | 0.9 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 2.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.6 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.4 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.6 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.3 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.4 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.3 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 0.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.8 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.4 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.3 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |