Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
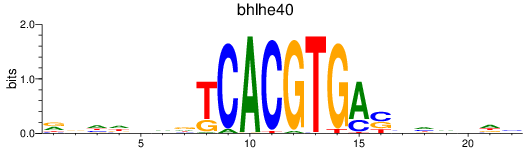
Results for bhlhe40
Z-value: 0.51
Transcription factors associated with bhlhe40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bhlhe40
|
ENSDARG00000004060 | basic helix-loop-helix family, member e40 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bhlhe40 | dr11_v1_chr11_-_35763323_35763323 | -0.99 | 4.0e-14 | Click! |
Activity profile of bhlhe40 motif
Sorted Z-values of bhlhe40 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_32622933 | 1.77 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr17_-_25331439 | 1.26 |
ENSDART00000155422
ENSDART00000082324 |
zpcx
|
zona pellucida protein C |
| chr9_-_34269066 | 1.20 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr19_+_791538 | 1.13 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr8_+_13700605 | 1.04 |
ENSDART00000144516
|
lonrf1l
|
LON peptidase N-terminal domain and ring finger 1, like |
| chr11_+_5499661 | 1.03 |
ENSDART00000027850
|
slc35e1
|
solute carrier family 35, member E1 |
| chr5_-_22130937 | 1.01 |
ENSDART00000138606
|
las1l
|
LAS1-like, ribosome biogenesis factor |
| chr12_+_19958845 | 1.00 |
ENSDART00000193248
|
ercc4
|
excision repair cross-complementation group 4 |
| chr6_-_49547680 | 0.94 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr5_+_41476443 | 0.89 |
ENSDART00000145228
ENSDART00000137981 ENSDART00000142538 |
pias2
|
protein inhibitor of activated STAT, 2 |
| chr10_+_32050906 | 0.88 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr7_+_51795667 | 0.85 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr6_+_149405 | 0.85 |
ENSDART00000161154
|
fdx1l
|
ferredoxin 1-like |
| chr10_+_5268054 | 0.83 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr25_+_3788074 | 0.80 |
ENSDART00000154008
|
chid1
|
chitinase domain containing 1 |
| chr9_-_10804796 | 0.79 |
ENSDART00000134911
|
si:ch1073-416j23.1
|
si:ch1073-416j23.1 |
| chr9_-_27868267 | 0.77 |
ENSDART00000079502
|
dbr1
|
debranching RNA lariats 1 |
| chr24_-_6024466 | 0.77 |
ENSDART00000040865
|
pdss1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr13_-_12389748 | 0.76 |
ENSDART00000141606
|
commd8
|
COMM domain containing 8 |
| chr17_+_30843881 | 0.75 |
ENSDART00000149600
ENSDART00000148547 |
tpp1
|
tripeptidyl peptidase I |
| chr19_+_15441022 | 0.75 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr19_+_9111550 | 0.74 |
ENSDART00000088336
|
setdb1a
|
SET domain, bifurcated 1a |
| chr14_-_16810401 | 0.73 |
ENSDART00000158396
ENSDART00000170758 |
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr4_-_9196291 | 0.73 |
ENSDART00000153963
|
hcfc2
|
host cell factor C2 |
| chr24_+_33462800 | 0.72 |
ENSDART00000166666
ENSDART00000050826 |
rmc1
|
regulator of MON1-CCZ1 |
| chr7_-_41881177 | 0.72 |
ENSDART00000174258
ENSDART00000018972 |
zgc:92818
|
zgc:92818 |
| chr5_+_26204561 | 0.70 |
ENSDART00000137178
|
marveld2b
|
MARVEL domain containing 2b |
| chr25_+_3788443 | 0.67 |
ENSDART00000189747
|
chid1
|
chitinase domain containing 1 |
| chr23_-_10786400 | 0.67 |
ENSDART00000055038
|
rybpa
|
RING1 and YY1 binding protein a |
| chr19_+_15440841 | 0.64 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr1_-_47114310 | 0.61 |
ENSDART00000144899
ENSDART00000053157 |
setd4
|
SET domain containing 4 |
| chr20_-_40766387 | 0.59 |
ENSDART00000061173
|
hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr9_+_16854121 | 0.59 |
ENSDART00000110866
|
cln5
|
CLN5, intracellular trafficking protein |
| chr16_+_33143503 | 0.59 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr25_-_19608382 | 0.58 |
ENSDART00000022279
ENSDART00000135201 ENSDART00000147223 ENSDART00000190220 ENSDART00000184242 ENSDART00000166824 |
gtse1
|
G-2 and S-phase expressed 1 |
| chr22_+_16759010 | 0.58 |
ENSDART00000079638
ENSDART00000113099 |
tm2d1
|
TM2 domain containing 1 |
| chr15_+_1534644 | 0.57 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr16_+_25296389 | 0.50 |
ENSDART00000114528
|
tbc1d31
|
TBC1 domain family, member 31 |
| chr23_-_2901167 | 0.50 |
ENSDART00000165955
ENSDART00000190616 |
zhx3
|
zinc fingers and homeoboxes 3 |
| chr24_-_42072886 | 0.48 |
ENSDART00000171389
|
CABZ01095370.1
|
|
| chr14_-_36345175 | 0.48 |
ENSDART00000077823
|
lrit3a
|
info leucine-rich repeat, immunoglobulin-like and transmembrane domains 3a |
| chr12_+_18529228 | 0.47 |
ENSDART00000193931
ENSDART00000146260 |
meiob
msrb1b
|
meiosis specific with OB domains methionine sulfoxide reductase B1b |
| chr15_-_44077937 | 0.47 |
ENSDART00000110112
|
lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr14_-_36437249 | 0.46 |
ENSDART00000016728
|
aga
|
aspartylglucosaminidase |
| chr17_+_25331576 | 0.46 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr15_+_34939906 | 0.46 |
ENSDART00000131182
|
si:ch73-95l15.3
|
si:ch73-95l15.3 |
| chr9_-_10805231 | 0.46 |
ENSDART00000193913
ENSDART00000078348 |
si:ch1073-416j23.1
|
si:ch1073-416j23.1 |
| chr23_+_32028574 | 0.44 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr22_-_16758973 | 0.43 |
ENSDART00000145208
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr20_-_45040916 | 0.41 |
ENSDART00000190001
|
klhl29
|
kelch-like family member 29 |
| chr10_-_24724388 | 0.40 |
ENSDART00000148582
|
smpd1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr23_-_22130778 | 0.39 |
ENSDART00000079212
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr4_+_17655872 | 0.39 |
ENSDART00000066999
|
washc3
|
WASH complex subunit 3 |
| chr8_-_25817106 | 0.37 |
ENSDART00000099364
|
taf10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr25_+_10547228 | 0.36 |
ENSDART00000067678
|
zgc:110339
|
zgc:110339 |
| chr21_+_43178831 | 0.35 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr16_+_19029297 | 0.33 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr20_+_13969414 | 0.33 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr21_-_4539899 | 0.32 |
ENSDART00000112460
|
dolk
|
dolichol kinase |
| chr17_-_15229787 | 0.31 |
ENSDART00000039165
|
styx
|
serine/threonine/tyrosine interacting protein |
| chr5_-_4532516 | 0.30 |
ENSDART00000192398
|
cst14b.1
|
cystatin 14b, tandem duplicate 1 |
| chr1_+_50639416 | 0.30 |
ENSDART00000141977
|
herc3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr19_+_40379771 | 0.29 |
ENSDART00000017917
ENSDART00000110699 |
vps50
vps50
|
VPS50 EARP/GARPII complex subunit VPS50 EARP/GARPII complex subunit |
| chr3_-_4501026 | 0.28 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr17_-_44440832 | 0.28 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr7_-_14446512 | 0.28 |
ENSDART00000041577
|
kif7
|
kinesin family member 7 |
| chr15_-_26570948 | 0.27 |
ENSDART00000156621
|
wdr81
|
WD repeat domain 81 |
| chr22_-_16759151 | 0.27 |
ENSDART00000191880
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr12_+_34891529 | 0.27 |
ENSDART00000015643
|
tbcc
|
tubulin folding cofactor C |
| chr23_+_35847200 | 0.26 |
ENSDART00000129222
|
rarga
|
retinoic acid receptor gamma a |
| chr1_+_17695426 | 0.23 |
ENSDART00000103236
|
ankrd37
|
ankyrin repeat domain 37 |
| chr14_-_45595711 | 0.23 |
ENSDART00000074038
|
scyl1
|
SCY1-like, kinase-like 1 |
| chr12_+_17933775 | 0.22 |
ENSDART00000186047
ENSDART00000160586 |
trrap
|
transformation/transcription domain-associated protein |
| chr20_-_29683754 | 0.21 |
ENSDART00000130599
ENSDART00000015928 ENSDART00000131219 |
si:ch211-195d17.2
|
si:ch211-195d17.2 |
| chr20_-_13140309 | 0.21 |
ENSDART00000020703
ENSDART00000188594 |
ints7
|
integrator complex subunit 7 |
| chr16_+_7985886 | 0.21 |
ENSDART00000126041
|
ano10a
|
anoctamin 10a |
| chr3_+_16922226 | 0.20 |
ENSDART00000017646
|
atp6v0a1a
|
ATPase H+ transporting V0 subunit a1a |
| chr3_-_25420931 | 0.20 |
ENSDART00000109601
ENSDART00000182184 |
bptf
|
bromodomain PHD finger transcription factor |
| chr12_-_7280551 | 0.18 |
ENSDART00000061633
|
zgc:171971
|
zgc:171971 |
| chr24_+_32472155 | 0.17 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr4_+_13696537 | 0.16 |
ENSDART00000109195
ENSDART00000122041 ENSDART00000192554 |
nrcama
|
neuronal cell adhesion molecule a |
| chr16_-_7228276 | 0.16 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr14_+_44804326 | 0.15 |
ENSDART00000079866
ENSDART00000172974 |
slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr5_-_9540641 | 0.15 |
ENSDART00000124384
ENSDART00000160079 |
gak
|
cyclin G associated kinase |
| chr15_-_41807371 | 0.15 |
ENSDART00000156819
|
slc25a36b
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36b |
| chr9_-_1200187 | 0.15 |
ENSDART00000158760
|
ino80da
|
INO80 complex subunit Da |
| chr13_+_40692804 | 0.14 |
ENSDART00000109822
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr13_+_12389849 | 0.14 |
ENSDART00000086525
|
atp10d
|
ATPase phospholipid transporting 10D |
| chr15_-_34418525 | 0.12 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr17_-_37299394 | 0.12 |
ENSDART00000154414
|
ptgr2
|
prostaglandin reductase 2 |
| chr13_+_7164345 | 0.11 |
ENSDART00000022051
|
gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr10_+_22890791 | 0.11 |
ENSDART00000176011
|
arrb2a
|
arrestin, beta 2a |
| chr7_+_1442059 | 0.10 |
ENSDART00000173391
|
si:cabz01090193.1
|
si:cabz01090193.1 |
| chr5_+_61839707 | 0.09 |
ENSDART00000163051
|
CR759879.1
|
Danio rerio interferon-induced protein with tetratricopeptide repeats 5 (LOC572297), mRNA. |
| chr18_+_31280984 | 0.09 |
ENSDART00000170285
ENSDART00000150608 ENSDART00000159720 |
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr20_-_211920 | 0.09 |
ENSDART00000104790
|
znf292b
|
zinc finger protein 292b |
| chr19_-_38830582 | 0.09 |
ENSDART00000189966
ENSDART00000183055 |
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr23_+_42131509 | 0.09 |
ENSDART00000184544
|
SHISAL2A
|
shisa like 2A |
| chr4_+_17353714 | 0.08 |
ENSDART00000136299
|
nup37
|
nucleoporin 37 |
| chr8_+_41229233 | 0.08 |
ENSDART00000131135
|
zgc:152830
|
zgc:152830 |
| chr2_-_32486080 | 0.07 |
ENSDART00000110821
|
ttc19
|
tetratricopeptide repeat domain 19 |
| chr14_-_3071564 | 0.07 |
ENSDART00000166136
ENSDART00000167138 |
slc35a4
|
solute carrier family 35, member A4 |
| chr22_+_8800432 | 0.06 |
ENSDART00000139940
|
si:dkey-182g1.6
|
si:dkey-182g1.6 |
| chr2_+_50967947 | 0.06 |
ENSDART00000162288
|
si:ch211-249o11.5
|
si:ch211-249o11.5 |
| chr24_+_3307857 | 0.06 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr4_+_13640903 | 0.05 |
ENSDART00000155349
|
pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr25_-_1323623 | 0.05 |
ENSDART00000156532
ENSDART00000157163 ENSDART00000156062 ENSDART00000082447 ENSDART00000189175 |
calml4b
|
calmodulin-like 4b |
| chr14_-_25452503 | 0.04 |
ENSDART00000148652
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr20_-_51386511 | 0.04 |
ENSDART00000151429
ENSDART00000066072 |
rnf8
|
ring finger protein 8, E3 ubiquitin protein ligase |
| chr21_-_14251306 | 0.04 |
ENSDART00000114715
ENSDART00000181380 |
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr11_-_44945636 | 0.04 |
ENSDART00000157658
|
orc2
|
origin recognition complex, subunit 2 |
| chr9_+_54686686 | 0.03 |
ENSDART00000066198
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr12_-_43438148 | 0.03 |
ENSDART00000004122
|
ptprea
|
protein tyrosine phosphatase, receptor type, E, a |
| chr3_+_12784460 | 0.03 |
ENSDART00000168382
|
cyp2k8
|
cytochrome P450, family 2, subfamily K, polypeptide 8 |
| chr3_-_25421504 | 0.03 |
ENSDART00000154200
|
bptf
|
bromodomain PHD finger transcription factor |
| chr4_-_17353972 | 0.02 |
ENSDART00000041529
|
parpbp
|
PARP1 binding protein |
| chr5_+_57210237 | 0.01 |
ENSDART00000167660
|
pja2
|
praja ring finger ubiquitin ligase 2 |
| chr18_+_39074139 | 0.01 |
ENSDART00000142390
|
gnb5b
|
guanine nucleotide binding protein (G protein), beta 5b |
| chr1_-_54765262 | 0.01 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr23_+_26079467 | 0.01 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr21_+_45816030 | 0.01 |
ENSDART00000187056
|
pitx1
|
paired-like homeodomain 1 |
| chr25_-_35599887 | 0.00 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
Network of associatons between targets according to the STRING database.
First level regulatory network of bhlhe40
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.3 | 0.3 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.1 | 0.3 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.5 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 0.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 1.2 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 0.9 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.7 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.2 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.2 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.8 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.8 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.4 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.9 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.0 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.2 | GO:0072318 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.6 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 1.8 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0035675 | neuromast hair cell development(GO:0035675) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.4 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.3 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.7 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 5.5 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.9 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 1.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 1.0 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.5 | GO:0033745 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.1 | 1.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 1.5 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 0.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.7 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.9 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.8 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.8 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.6 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.0 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 1.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.0 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |