Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
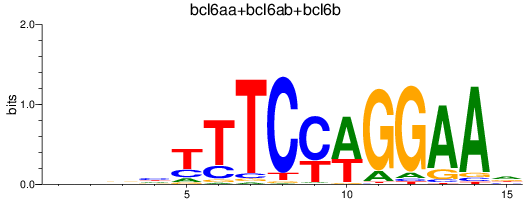
Results for bcl6aa+bcl6ab+bcl6b
Z-value: 0.28
Transcription factors associated with bcl6aa+bcl6ab+bcl6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bcl6ab
|
ENSDARG00000069295 | BCL6A transcription repressor b |
|
bcl6b
|
ENSDARG00000069335 | BCL6B transcription repressor |
|
bcl6aa
|
ENSDARG00000070864 | BCL6A transcription repressor a |
|
bcl6aa
|
ENSDARG00000111395 | BCL6A transcription repressor a |
|
bcl6aa
|
ENSDARG00000112502 | BCL6A transcription repressor a |
|
bcl6aa
|
ENSDARG00000116260 | BCL6A transcription repressor a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bcl6b | dr11_v1_chr7_+_20017211_20017211 | 0.94 | 1.1e-08 | Click! |
| bcl6ab | dr11_v1_chr2_-_10098191_10098191 | 0.74 | 4.3e-04 | Click! |
| bcl6a | dr11_v1_chr6_-_28222592_28222592 | -0.62 | 5.8e-03 | Click! |
Activity profile of bcl6aa+bcl6ab+bcl6b motif
Sorted Z-values of bcl6aa+bcl6ab+bcl6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_17964525 | 1.33 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr10_+_40324395 | 1.19 |
ENSDART00000147205
|
gltpb
|
glycolipid transfer protein b |
| chr6_-_40352215 | 0.94 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr24_+_31361407 | 0.91 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr17_+_7522777 | 0.89 |
ENSDART00000184723
|
klhl10b.2
|
kelch-like family member 10b, tandem duplicate 2 |
| chr12_-_27242498 | 0.82 |
ENSDART00000152609
ENSDART00000152170 |
si:dkey-11c5.11
|
si:dkey-11c5.11 |
| chr16_-_54465271 | 0.82 |
ENSDART00000185978
|
cyp11c1
|
cytochrome P450, family 11, subfamily C, polypeptide 1 |
| chr14_-_24397718 | 0.80 |
ENSDART00000110164
|
si:ch211-163m17.4
|
si:ch211-163m17.4 |
| chr13_-_11967769 | 0.74 |
ENSDART00000158369
|
ARL3 (1 of many)
|
zgc:110197 |
| chr9_+_40825065 | 0.74 |
ENSDART00000137673
|
si:dkey-95p16.2
|
si:dkey-95p16.2 |
| chr22_-_3344613 | 0.72 |
ENSDART00000165600
|
tbxa2r
|
thromboxane A2 receptor |
| chr20_+_39457598 | 0.72 |
ENSDART00000140931
ENSDART00000156176 |
pimr128
|
Pim proto-oncogene, serine/threonine kinase, related 128 |
| chr21_+_35215810 | 0.70 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr23_-_35483163 | 0.66 |
ENSDART00000138660
ENSDART00000113643 ENSDART00000189269 |
fbxo25
|
F-box protein 25 |
| chr20_-_43533096 | 0.61 |
ENSDART00000145199
|
pimr134
|
Pim proto-oncogene, serine/threonine kinase, related 134 |
| chr13_-_37608441 | 0.60 |
ENSDART00000140230
|
zgc:123068
|
zgc:123068 |
| chr8_-_23783633 | 0.58 |
ENSDART00000132657
|
si:ch211-163l21.7
|
si:ch211-163l21.7 |
| chr13_-_37642890 | 0.54 |
ENSDART00000146483
ENSDART00000136071 ENSDART00000111786 |
si:dkey-188i13.9
|
si:dkey-188i13.9 |
| chr18_+_25752592 | 0.53 |
ENSDART00000111767
|
si:ch211-39k3.2
|
si:ch211-39k3.2 |
| chr6_+_48041759 | 0.53 |
ENSDART00000140086
|
si:dkey-92f12.2
|
si:dkey-92f12.2 |
| chr20_+_23440632 | 0.49 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr4_-_8152746 | 0.48 |
ENSDART00000012928
ENSDART00000177482 |
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr2_-_10703621 | 0.47 |
ENSDART00000005944
|
rpl5a
|
ribosomal protein L5a |
| chr9_+_26103814 | 0.46 |
ENSDART00000026011
|
efnb2a
|
ephrin-B2a |
| chr4_+_19127973 | 0.46 |
ENSDART00000136611
|
si:dkey-21o22.2
|
si:dkey-21o22.2 |
| chr21_+_26720803 | 0.44 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr14_-_21123325 | 0.43 |
ENSDART00000180889
ENSDART00000188539 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr3_-_60602432 | 0.42 |
ENSDART00000163235
|
srsf2b
|
serine/arginine-rich splicing factor 2b |
| chr22_+_7480465 | 0.42 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr17_-_24564674 | 0.41 |
ENSDART00000105435
ENSDART00000135086 |
abch1
|
ATP-binding cassette, sub-family H, member 1 |
| chr10_-_3332362 | 0.39 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr16_-_41004731 | 0.39 |
ENSDART00000102591
|
KCNV1
|
si:dkey-201i6.2 |
| chr6_+_12865137 | 0.38 |
ENSDART00000090065
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr22_-_16180467 | 0.38 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr7_+_32369026 | 0.38 |
ENSDART00000169588
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr3_-_55139127 | 0.38 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr14_+_31651533 | 0.37 |
ENSDART00000172835
|
fhl1a
|
four and a half LIM domains 1a |
| chr22_-_37611681 | 0.37 |
ENSDART00000028085
|
ttc14
|
tetratricopeptide repeat domain 14 |
| chr20_-_43495506 | 0.36 |
ENSDART00000170679
|
pimr132
|
Pim proto-oncogene, serine/threonine kinase, related 132 |
| chr4_+_12342173 | 0.36 |
ENSDART00000161518
|
pimr214
|
Pim proto-oncogene, serine/threonine kinase, related 214 |
| chr7_+_38509333 | 0.36 |
ENSDART00000153482
|
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr3_-_31655796 | 0.36 |
ENSDART00000181052
|
CU062431.1
|
|
| chr1_-_9114936 | 0.35 |
ENSDART00000163624
ENSDART00000132910 |
si:ch211-14k19.8
vap
|
si:ch211-14k19.8 vascular associated protein |
| chr7_+_61876641 | 0.35 |
ENSDART00000165409
|
CR382383.2
|
|
| chr4_-_12323228 | 0.34 |
ENSDART00000081089
|
il17ra1a
|
interleukin 17 receptor A1a |
| chr21_+_17956024 | 0.33 |
ENSDART00000142468
|
dnai1.2
|
dynein, axonemal, intermediate chain 1, paralog 2 |
| chr22_+_37902976 | 0.32 |
ENSDART00000169327
|
kcnh1a
|
potassium voltage-gated channel, subfamily H (eag-related), member 1a |
| chr8_-_39654669 | 0.32 |
ENSDART00000145677
|
si:dkey-63d15.12
|
si:dkey-63d15.12 |
| chr11_+_30321116 | 0.32 |
ENSDART00000187921
ENSDART00000127075 |
ugt1b1
|
UDP glucuronosyltransferase 1 family, polypeptide B1 |
| chr14_+_11457500 | 0.32 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr25_+_25438165 | 0.30 |
ENSDART00000161369
|
LRRC56
|
si:ch211-103e16.5 |
| chr19_-_23892243 | 0.30 |
ENSDART00000132384
|
si:dkey-222b8.4
|
si:dkey-222b8.4 |
| chr6_-_35446110 | 0.29 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr23_-_19715799 | 0.29 |
ENSDART00000142072
ENSDART00000032744 ENSDART00000131860 |
rpl10
|
ribosomal protein L10 |
| chr23_-_14830627 | 0.29 |
ENSDART00000134659
|
sla2
|
Src-like-adaptor 2 |
| chr3_-_55121125 | 0.28 |
ENSDART00000125092
|
hbae1
|
hemoglobin, alpha embryonic 1 |
| chr4_+_12367546 | 0.28 |
ENSDART00000161860
|
pimr169
|
Pim proto-oncogene, serine/threonine kinase, related 169 |
| chr3_-_13461056 | 0.27 |
ENSDART00000137678
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr5_+_13359146 | 0.27 |
ENSDART00000179839
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr5_-_32882162 | 0.27 |
ENSDART00000085769
|
lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr21_-_22715297 | 0.27 |
ENSDART00000065548
|
c1qb
|
complement component 1, q subcomponent, B chain |
| chr12_+_7491690 | 0.27 |
ENSDART00000152564
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr1_+_30083578 | 0.27 |
ENSDART00000101779
ENSDART00000143639 |
vps8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr13_+_6092135 | 0.27 |
ENSDART00000162738
|
fam120b
|
family with sequence similarity 120B |
| chr16_-_45917322 | 0.27 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr24_-_37472727 | 0.27 |
ENSDART00000134152
|
cluap1
|
clusterin associated protein 1 |
| chr6_+_43015916 | 0.26 |
ENSDART00000064888
|
tcta
|
T cell leukemia translocation altered |
| chr19_-_7240627 | 0.25 |
ENSDART00000181874
|
FP101875.2
|
|
| chr24_+_9590188 | 0.25 |
ENSDART00000137092
|
si:dkey-96n2.1
|
si:dkey-96n2.1 |
| chr7_+_52766211 | 0.25 |
ENSDART00000186191
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr15_-_23793641 | 0.25 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr21_-_2238277 | 0.24 |
ENSDART00000165754
|
si:dkey-50i6.6
|
si:dkey-50i6.6 |
| chr2_+_3428357 | 0.24 |
ENSDART00000125967
|
CU633991.1
|
|
| chr4_-_18436899 | 0.24 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr14_-_5678457 | 0.24 |
ENSDART00000012116
|
tlx2
|
T cell leukemia homeobox 2 |
| chr7_+_26029672 | 0.24 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr6_+_9881093 | 0.24 |
ENSDART00000151558
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr5_+_63668735 | 0.23 |
ENSDART00000134261
ENSDART00000097330 |
dnm1b
|
dynamin 1b |
| chr13_-_47554194 | 0.23 |
ENSDART00000181508
|
acoxl
|
acyl-CoA oxidase-like |
| chr5_+_25072894 | 0.23 |
ENSDART00000012268
|
mrpl41
|
mitochondrial ribosomal protein L41 |
| chr14_+_24241241 | 0.23 |
ENSDART00000022377
|
nkx2.5
|
NK2 homeobox 5 |
| chr3_-_52661242 | 0.23 |
ENSDART00000138018
|
zgc:113210
|
zgc:113210 |
| chr3_-_13461361 | 0.23 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr23_+_19670085 | 0.22 |
ENSDART00000031872
|
kctd6b
|
potassium channel tetramerization domain containing 6b |
| chr19_+_5418006 | 0.22 |
ENSDART00000132874
|
eif1b
|
eukaryotic translation initiation factor 1B |
| chr18_-_16801033 | 0.22 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr5_+_4366431 | 0.22 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr14_-_38828057 | 0.21 |
ENSDART00000186088
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr9_+_23748342 | 0.20 |
ENSDART00000019053
|
faima
|
Fas apoptotic inhibitory molecule a |
| chr4_+_359970 | 0.20 |
ENSDART00000139832
|
tmem181
|
transmembrane protein 181 |
| chr1_-_26398361 | 0.20 |
ENSDART00000160183
|
ppa2
|
pyrophosphatase (inorganic) 2 |
| chr16_+_46294337 | 0.20 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr20_-_47188966 | 0.19 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr3_-_55128258 | 0.19 |
ENSDART00000101734
|
hbae1
|
hemoglobin, alpha embryonic 1 |
| chr7_+_25053331 | 0.19 |
ENSDART00000173998
|
si:dkey-23i12.7
|
si:dkey-23i12.7 |
| chr21_-_27338639 | 0.19 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr14_+_6159356 | 0.19 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr3_-_44059902 | 0.19 |
ENSDART00000158485
ENSDART00000159088 ENSDART00000165628 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr3_-_60316118 | 0.18 |
ENSDART00000171458
|
si:ch211-214b16.2
|
si:ch211-214b16.2 |
| chr3_+_32492467 | 0.18 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr4_+_47257854 | 0.18 |
ENSDART00000173868
|
crestin
|
crestin |
| chr6_+_11989537 | 0.18 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr19_+_25504645 | 0.18 |
ENSDART00000143292
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr16_-_36834505 | 0.18 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr13_-_8888334 | 0.18 |
ENSDART00000059881
|
plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr6_-_35106425 | 0.18 |
ENSDART00000165139
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr3_-_7147226 | 0.18 |
ENSDART00000178155
|
BX005085.5
|
|
| chr7_-_22941472 | 0.18 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr5_+_24179307 | 0.18 |
ENSDART00000051552
|
mpdu1a
|
mannose-P-dolichol utilization defect 1a |
| chr3_-_44012748 | 0.17 |
ENSDART00000167248
ENSDART00000157463 ENSDART00000159111 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr20_+_53521648 | 0.17 |
ENSDART00000139794
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr15_-_2010926 | 0.17 |
ENSDART00000155688
|
dock10
|
dedicator of cytokinesis 10 |
| chr8_+_30671060 | 0.17 |
ENSDART00000193749
|
adora2aa
|
adenosine A2a receptor a |
| chr18_-_14860435 | 0.17 |
ENSDART00000018502
|
mapk12a
|
mitogen-activated protein kinase 12a |
| chr7_+_32369463 | 0.17 |
ENSDART00000180544
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr22_-_15578402 | 0.17 |
ENSDART00000062986
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr10_-_8358396 | 0.17 |
ENSDART00000059322
|
csgalnact1a
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1a |
| chr19_+_32856907 | 0.17 |
ENSDART00000148232
|
rpl30
|
ribosomal protein L30 |
| chr21_-_2217685 | 0.17 |
ENSDART00000159315
|
si:dkey-50i6.5
|
si:dkey-50i6.5 |
| chr9_-_23922011 | 0.17 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr18_+_50907675 | 0.16 |
ENSDART00000159950
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr9_+_2002701 | 0.16 |
ENSDART00000082329
|
evx2
|
even-skipped homeobox 2 |
| chr8_-_41228530 | 0.16 |
ENSDART00000165949
ENSDART00000173055 |
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr7_+_50448478 | 0.16 |
ENSDART00000098815
|
serinc4
|
serine incorporator 4 |
| chr24_+_11381400 | 0.16 |
ENSDART00000058703
|
ackr4b
|
atypical chemokine receptor 4b |
| chr9_-_21912227 | 0.16 |
ENSDART00000145576
|
lmo7a
|
LIM domain 7a |
| chr22_+_1462177 | 0.16 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr2_+_327081 | 0.16 |
ENSDART00000155595
|
zgc:174263
|
zgc:174263 |
| chr4_-_67969695 | 0.16 |
ENSDART00000190016
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr15_-_3282220 | 0.16 |
ENSDART00000092942
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr20_+_52458765 | 0.15 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr3_+_62236443 | 0.15 |
ENSDART00000180482
ENSDART00000193718 |
BX470259.4
|
|
| chr11_-_38914265 | 0.15 |
ENSDART00000141229
|
si:ch211-122l14.4
|
si:ch211-122l14.4 |
| chr15_-_40246396 | 0.15 |
ENSDART00000063777
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr19_-_41405187 | 0.15 |
ENSDART00000038038
ENSDART00000172350 |
sem1
|
SEM1, 26S proteasome complex subunit |
| chr19_-_35428815 | 0.15 |
ENSDART00000169006
ENSDART00000003167 |
ak2
|
adenylate kinase 2 |
| chr14_-_28567845 | 0.15 |
ENSDART00000126095
|
insb
|
preproinsulin b |
| chr14_-_24410673 | 0.15 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr17_+_10318071 | 0.14 |
ENSDART00000161844
|
foxa1
|
forkhead box A1 |
| chr1_+_10318089 | 0.14 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr20_+_25552057 | 0.14 |
ENSDART00000102913
|
cyp2v1
|
cytochrome P450, family 2, subfamily V, polypeptide 1 |
| chr10_+_21722892 | 0.14 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr9_-_6372535 | 0.14 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr8_-_19246342 | 0.14 |
ENSDART00000147172
|
abhd17ab
|
abhydrolase domain containing 17Ab |
| chr20_-_23440955 | 0.14 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr17_-_48743076 | 0.13 |
ENSDART00000173117
|
kcnk17
|
potassium channel, subfamily K, member 17 |
| chr9_+_6934491 | 0.13 |
ENSDART00000114323
|
mfsd9
|
major facilitator superfamily domain containing 9 |
| chr10_-_4375190 | 0.13 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr15_+_23550890 | 0.13 |
ENSDART00000009796
ENSDART00000152720 |
MARK4
|
si:dkey-31m14.7 |
| chr5_-_25582721 | 0.13 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr24_-_33291784 | 0.13 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr21_-_18982337 | 0.13 |
ENSDART00000147246
|
si:ch211-222n4.6
|
si:ch211-222n4.6 |
| chr1_-_58664854 | 0.13 |
ENSDART00000109528
|
adgre5b.3
|
adhesion G protein-coupled receptor E5b, duplicate 3 |
| chr19_-_6134802 | 0.13 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr5_-_42872712 | 0.13 |
ENSDART00000003947
|
flot2a
|
flotillin 2a |
| chr16_+_25343934 | 0.13 |
ENSDART00000109710
|
zfat
|
zinc finger and AT hook domain containing |
| chr22_+_3238474 | 0.13 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr21_+_45626136 | 0.13 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr1_+_49673489 | 0.12 |
ENSDART00000135487
|
tsga10
|
testis specific, 10 |
| chr20_-_38446891 | 0.12 |
ENSDART00000192013
|
itpkb
|
inositol-trisphosphate 3-kinase B |
| chr20_-_39391833 | 0.12 |
ENSDART00000135149
|
si:dkey-217m5.8
|
si:dkey-217m5.8 |
| chr6_-_8498908 | 0.12 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr13_-_37631092 | 0.12 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr11_-_36350876 | 0.12 |
ENSDART00000146495
ENSDART00000020655 |
psma5
|
proteasome subunit alpha 5 |
| chr4_+_74141400 | 0.12 |
ENSDART00000166994
|
trim24
|
tripartite motif containing 24 |
| chr10_+_21730585 | 0.12 |
ENSDART00000188576
|
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr9_-_54304684 | 0.12 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr21_+_26073104 | 0.12 |
ENSDART00000193273
|
rpl23a
|
ribosomal protein L23a |
| chr13_-_49819027 | 0.11 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr24_+_10397865 | 0.11 |
ENSDART00000155557
|
si:ch211-69l10.4
|
si:ch211-69l10.4 |
| chr22_-_27706576 | 0.11 |
ENSDART00000188564
|
CR547131.1
|
|
| chr1_-_9195629 | 0.11 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr21_-_2248239 | 0.11 |
ENSDART00000169448
|
si:ch73-299h12.1
|
si:ch73-299h12.1 |
| chr18_+_13315739 | 0.11 |
ENSDART00000143404
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr19_-_38611814 | 0.11 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr23_+_42292748 | 0.10 |
ENSDART00000166113
ENSDART00000158684 |
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr24_-_21991770 | 0.10 |
ENSDART00000147296
|
si:rp71-1f1.4
|
si:rp71-1f1.4 |
| chr22_+_34691020 | 0.10 |
ENSDART00000192202
|
hykk.1
|
hydroxylysine kinase, tandem duplicate 1 |
| chr16_+_25809235 | 0.10 |
ENSDART00000181196
|
irgq1
|
immunity-related GTPase family, q1 |
| chrM_+_4993 | 0.10 |
ENSDART00000093600
|
mt-nd2
|
NADH dehydrogenase 2, mitochondrial |
| chr18_+_18863167 | 0.10 |
ENSDART00000091094
|
pllp
|
plasmolipin |
| chr9_+_29585943 | 0.10 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr19_-_5351980 | 0.10 |
ENSDART00000163304
ENSDART00000027701 |
krt92
|
keratin 92 |
| chr23_-_19682971 | 0.10 |
ENSDART00000048891
|
zgc:193598
|
zgc:193598 |
| chr3_+_19299309 | 0.10 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr4_+_5642696 | 0.10 |
ENSDART00000028941
|
mrps18a
|
mitochondrial ribosomal protein S18A |
| chr4_-_13548806 | 0.09 |
ENSDART00000067155
|
il22
|
interleukin 22 |
| chr9_-_370225 | 0.09 |
ENSDART00000163637
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr17_-_608857 | 0.09 |
ENSDART00000163431
|
KLHL28
|
kelch like family member 28 |
| chr24_-_37877743 | 0.09 |
ENSDART00000105658
|
tmem204
|
transmembrane protein 204 |
| chr18_+_37259800 | 0.09 |
ENSDART00000078053
ENSDART00000132577 |
tbcb
|
tubulin folding cofactor B |
| chr5_-_55600689 | 0.09 |
ENSDART00000013229
|
gnaq
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr24_+_7828097 | 0.09 |
ENSDART00000134975
|
zgc:101569
|
zgc:101569 |
| chr13_+_33688474 | 0.09 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr19_-_35229336 | 0.09 |
ENSDART00000054274
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr6_+_32834007 | 0.09 |
ENSDART00000157353
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr9_-_32813177 | 0.09 |
ENSDART00000012694
|
mxc
|
myxovirus (influenza virus) resistance C |
| chr19_-_3742472 | 0.08 |
ENSDART00000162132
|
btr22
|
bloodthirsty-related gene family, member 22 |
| chr11_-_22303678 | 0.08 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr10_+_22134606 | 0.08 |
ENSDART00000155228
|
si:dkey-4c2.11
|
si:dkey-4c2.11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of bcl6aa+bcl6ab+bcl6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 0.8 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.3 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.5 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.6 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 0.4 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.1 | 0.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 0.2 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.1 | 0.7 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.1 | 0.2 | GO:0098581 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.1 | 0.2 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.2 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.4 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.4 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.5 | GO:1901381 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.4 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 0.1 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.1 | GO:0033034 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 0.1 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 0.9 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.1 | GO:0019628 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.0 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0071887 | leukocyte apoptotic process(GO:0071887) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.1 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.0 | 0.2 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.5 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.0 | GO:0002839 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 1.1 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.0 | GO:0055026 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.0 | 0.3 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) syncytium formation(GO:0006949) |
| 0.0 | 0.1 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.1 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) regulation of mitophagy(GO:1903146) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0050688 | regulation of response to biotic stimulus(GO:0002831) regulation of defense response to virus(GO:0050688) |
| 0.0 | 0.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.9 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.0 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.2 | 1.2 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.7 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.1 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.2 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.2 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 0.1 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |