Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
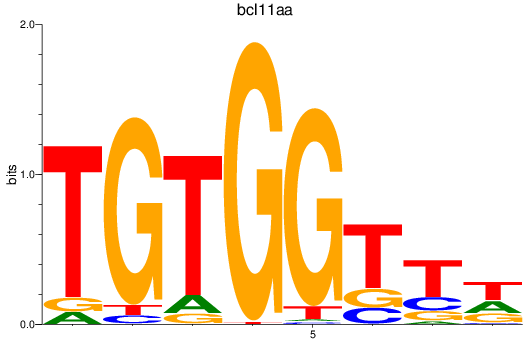
Results for bcl11aa
Z-value: 0.52
Transcription factors associated with bcl11aa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bcl11aa
|
ENSDARG00000061352 | BAF chromatin remodeling complex subunit BCL11A a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bcl11aa | dr11_v1_chr13_+_25846528_25846528 | -0.47 | 5.0e-02 | Click! |
Activity profile of bcl11aa motif
Sorted Z-values of bcl11aa motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_11923701 | 0.49 |
ENSDART00000109292
|
ubap2a
|
ubiquitin associated protein 2a |
| chr9_+_38457806 | 0.45 |
ENSDART00000142512
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr23_+_30898013 | 0.45 |
ENSDART00000146859
|
cables2a
|
Cdk5 and Abl enzyme substrate 2a |
| chr9_+_38458193 | 0.44 |
ENSDART00000008053
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr8_+_44703864 | 0.43 |
ENSDART00000016225
|
star
|
steroidogenic acute regulatory protein |
| chr6_-_16394528 | 0.42 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr14_+_24845941 | 0.42 |
ENSDART00000187513
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr25_+_4541211 | 0.38 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr25_+_18953756 | 0.37 |
ENSDART00000154291
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr19_-_11425542 | 0.36 |
ENSDART00000177875
ENSDART00000080762 |
sept7a
|
septin 7a |
| chr6_+_38626926 | 0.33 |
ENSDART00000190339
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr20_-_18789543 | 0.33 |
ENSDART00000182240
|
ccm2
|
cerebral cavernous malformation 2 |
| chr7_-_26263183 | 0.33 |
ENSDART00000079357
ENSDART00000190369 ENSDART00000193154 ENSDART00000101109 |
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr7_-_26262978 | 0.33 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr3_-_31833266 | 0.32 |
ENSDART00000084932
ENSDART00000183617 |
map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr25_-_14637660 | 0.32 |
ENSDART00000143666
|
nav2b
|
neuron navigator 2b |
| chr2_+_3516913 | 0.31 |
ENSDART00000109346
|
CU693445.1
|
|
| chr24_+_31361407 | 0.31 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr2_+_36046126 | 0.31 |
ENSDART00000085976
ENSDART00000171680 |
smg7
|
SMG7 nonsense mediated mRNA decay factor |
| chr13_+_11829072 | 0.29 |
ENSDART00000079356
ENSDART00000170160 |
sufu
|
suppressor of fused homolog (Drosophila) |
| chr2_+_21855036 | 0.29 |
ENSDART00000140012
|
ca8
|
carbonic anhydrase VIII |
| chr5_-_38384289 | 0.29 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr18_+_27571448 | 0.28 |
ENSDART00000147886
|
cd82b
|
CD82 molecule b |
| chr16_+_10963602 | 0.27 |
ENSDART00000141032
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr24_+_39277043 | 0.27 |
ENSDART00000165458
|
MPRIP
|
si:ch73-103b11.2 |
| chr11_+_18216404 | 0.27 |
ENSDART00000086437
|
tmcc1b
|
transmembrane and coiled-coil domain family 1b |
| chr4_+_23117557 | 0.27 |
ENSDART00000066909
|
slc35e3
|
solute carrier family 35, member E3 |
| chr11_+_4026229 | 0.27 |
ENSDART00000041417
|
camk1b
|
calcium/calmodulin-dependent protein kinase Ib |
| chr8_+_23381892 | 0.26 |
ENSDART00000180950
ENSDART00000063010 ENSDART00000074241 ENSDART00000142783 |
mapre1a
|
microtubule-associated protein, RP/EB family, member 1a |
| chr19_+_29798064 | 0.26 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr25_+_18954189 | 0.25 |
ENSDART00000123207
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr7_+_39706004 | 0.25 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr19_-_20403318 | 0.25 |
ENSDART00000136826
|
dazl
|
deleted in azoospermia-like |
| chr16_+_28547157 | 0.24 |
ENSDART00000109450
ENSDART00000165687 |
fam171a1
|
family with sequence similarity 171, member A1 |
| chr9_+_30112423 | 0.24 |
ENSDART00000112398
ENSDART00000013591 |
tfg
|
trk-fused gene |
| chr11_-_12801157 | 0.24 |
ENSDART00000103449
|
txlng
|
taxilin gamma |
| chr9_+_33267211 | 0.24 |
ENSDART00000025635
|
usp9
|
ubiquitin specific peptidase 9 |
| chr11_-_12800945 | 0.23 |
ENSDART00000191178
|
txlng
|
taxilin gamma |
| chr8_-_43456025 | 0.23 |
ENSDART00000001092
ENSDART00000140618 |
ncor2
|
nuclear receptor corepressor 2 |
| chr23_+_21978816 | 0.23 |
ENSDART00000087110
|
eif4g3b
|
eukaryotic translation initiation factor 4 gamma, 3b |
| chr15_+_39977461 | 0.23 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr9_+_51655636 | 0.23 |
ENSDART00000169908
|
RBMS1 (1 of many)
|
RNA binding motif single stranded interacting protein 1 |
| chr22_-_31517300 | 0.22 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr17_-_10043273 | 0.22 |
ENSDART00000156078
|
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr5_-_43682930 | 0.22 |
ENSDART00000075017
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr2_+_21855291 | 0.21 |
ENSDART00000186204
|
ca8
|
carbonic anhydrase VIII |
| chr7_+_59677273 | 0.20 |
ENSDART00000039535
ENSDART00000132044 |
trmt44
|
tRNA methyltransferase 44 homolog |
| chr16_-_13612650 | 0.19 |
ENSDART00000080372
|
dbpb
|
D site albumin promoter binding protein b |
| chr2_+_22694382 | 0.19 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr16_+_19014886 | 0.19 |
ENSDART00000079298
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr7_-_30553588 | 0.19 |
ENSDART00000139546
|
sltm
|
SAFB-like, transcription modulator |
| chr22_-_20309283 | 0.18 |
ENSDART00000182125
ENSDART00000048775 |
si:dkey-110c1.10
|
si:dkey-110c1.10 |
| chr22_-_10397600 | 0.18 |
ENSDART00000181964
ENSDART00000142886 |
nisch
|
nischarin |
| chr8_+_45294767 | 0.17 |
ENSDART00000191527
|
ubap2b
|
ubiquitin associated protein 2b |
| chr3_+_54761569 | 0.16 |
ENSDART00000135913
ENSDART00000180983 |
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr25_+_15997957 | 0.16 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr5_+_61556172 | 0.16 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr2_-_21786826 | 0.15 |
ENSDART00000016208
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr3_+_22335030 | 0.15 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr5_+_28313824 | 0.15 |
ENSDART00000135878
|
si:dkeyp-86b9.1
|
si:dkeyp-86b9.1 |
| chr4_-_19742300 | 0.15 |
ENSDART00000066964
ENSDART00000100952 |
hgfa
|
hepatocyte growth factor a |
| chr8_+_10835456 | 0.15 |
ENSDART00000151388
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr25_+_30238021 | 0.15 |
ENSDART00000123220
|
alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr23_+_21978584 | 0.15 |
ENSDART00000145172
|
eif4g3b
|
eukaryotic translation initiation factor 4 gamma, 3b |
| chr16_-_22863603 | 0.14 |
ENSDART00000098400
ENSDART00000183963 |
shc1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr2_+_48288461 | 0.14 |
ENSDART00000141495
|
hes6
|
hes family bHLH transcription factor 6 |
| chr12_-_19279103 | 0.14 |
ENSDART00000186669
|
si:ch211-141o9.10
|
si:ch211-141o9.10 |
| chr5_+_24156170 | 0.14 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr3_+_28576173 | 0.14 |
ENSDART00000151189
|
sept12
|
septin 12 |
| chr16_-_22863348 | 0.14 |
ENSDART00000147609
|
shc1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr5_-_38122126 | 0.14 |
ENSDART00000141791
ENSDART00000170528 |
si:ch211-284e13.6
|
si:ch211-284e13.6 |
| chr25_+_31929325 | 0.13 |
ENSDART00000181095
|
apba2a
|
amyloid beta (A4) precursor protein-binding, family A, member 2a |
| chr7_+_46368520 | 0.13 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr2_+_24786765 | 0.13 |
ENSDART00000141030
|
pde4ca
|
phosphodiesterase 4C, cAMP-specific a |
| chr19_+_19786117 | 0.13 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr5_-_41933912 | 0.12 |
ENSDART00000097574
|
ncor1
|
nuclear receptor corepressor 1 |
| chr4_+_16710001 | 0.12 |
ENSDART00000035899
|
pkp2
|
plakophilin 2 |
| chr24_-_21921262 | 0.12 |
ENSDART00000186061
ENSDART00000187846 |
tagln3b
|
transgelin 3b |
| chr2_-_14387335 | 0.12 |
ENSDART00000189332
ENSDART00000164786 ENSDART00000188572 |
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr5_-_63425428 | 0.12 |
ENSDART00000163246
|
cntrl
|
centriolin |
| chr5_+_13359146 | 0.12 |
ENSDART00000179839
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr9_-_31108285 | 0.12 |
ENSDART00000003193
|
gpr183a
|
G protein-coupled receptor 183a |
| chr13_-_6081803 | 0.11 |
ENSDART00000099224
|
dld
|
deltaD |
| chr22_-_17729778 | 0.11 |
ENSDART00000192132
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr2_-_24369087 | 0.11 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr12_+_28856151 | 0.11 |
ENSDART00000152969
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr13_-_3324764 | 0.10 |
ENSDART00000102748
ENSDART00000114040 |
ubr2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr16_+_50741154 | 0.10 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr7_+_56735195 | 0.10 |
ENSDART00000082830
|
KIAA0895L
|
KIAA0895 like |
| chr18_+_19131773 | 0.10 |
ENSDART00000060766
|
rab11a
|
RAB11a, member RAS oncogene family |
| chr6_+_45918981 | 0.10 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr18_+_35742838 | 0.10 |
ENSDART00000088504
ENSDART00000140386 |
rasgrp4
|
RAS guanyl releasing protein 4 |
| chr23_-_18567088 | 0.09 |
ENSDART00000192371
|
sephs2
|
selenophosphate synthetase 2 |
| chr3_+_22905341 | 0.09 |
ENSDART00000111435
|
hdac5
|
histone deacetylase 5 |
| chr19_-_34011340 | 0.09 |
ENSDART00000172618
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr4_+_20255160 | 0.09 |
ENSDART00000188658
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr22_+_22438783 | 0.09 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr20_+_34543365 | 0.09 |
ENSDART00000152073
|
si:ch211-242b18.1
|
si:ch211-242b18.1 |
| chr4_+_12031958 | 0.08 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr19_-_7144548 | 0.08 |
ENSDART00000147177
ENSDART00000134850 |
psmb8a
psmb13a
|
proteasome subunit beta 8A proteasome subunit beta 13a |
| chr1_+_33217911 | 0.08 |
ENSDART00000136761
|
prkx
|
protein kinase, X-linked |
| chr14_-_30967284 | 0.08 |
ENSDART00000149435
|
il2rgb
|
interleukin 2 receptor, gamma b |
| chr11_-_11965033 | 0.07 |
ENSDART00000193683
|
abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr24_-_18877118 | 0.07 |
ENSDART00000092783
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr17_-_8899323 | 0.07 |
ENSDART00000081590
|
nkl.1
|
NK-lysin tandem duplicate 1 |
| chr16_+_27957808 | 0.07 |
ENSDART00000133696
|
znf804b
|
zinc finger protein 804B |
| chr6_-_21830405 | 0.07 |
ENSDART00000151803
ENSDART00000113497 |
setd5
|
SET domain containing 5 |
| chr19_+_7552699 | 0.07 |
ENSDART00000180788
ENSDART00000115058 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr13_+_24834199 | 0.07 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr15_+_37412883 | 0.07 |
ENSDART00000156474
|
zbtb32
|
zinc finger and BTB domain containing 32 |
| chr17_-_45254585 | 0.06 |
ENSDART00000185507
ENSDART00000172080 |
ttbk2a
|
tau tubulin kinase 2a |
| chr4_+_5215950 | 0.06 |
ENSDART00000140613
|
galnt8b.2
|
polypeptide N-acetylgalactosaminyltransferase 8b, tandem duplicate 2 |
| chr11_-_40647190 | 0.06 |
ENSDART00000173217
ENSDART00000173276 ENSDART00000147264 |
fam213b
|
family with sequence similarity 213, member B |
| chr25_-_22889519 | 0.05 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr25_-_35996141 | 0.05 |
ENSDART00000149074
|
sall1b
|
spalt-like transcription factor 1b |
| chr17_+_26569601 | 0.05 |
ENSDART00000153897
|
ndnfl
|
neuron-derived neurotrophic factor , like |
| chr24_-_18876877 | 0.05 |
ENSDART00000186269
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr18_-_2639351 | 0.05 |
ENSDART00000168106
|
relt
|
RELT, TNF receptor |
| chr15_-_34567370 | 0.05 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr17_+_23770848 | 0.05 |
ENSDART00000079646
|
kcnk18
|
potassium channel, subfamily K, member 18 |
| chr16_+_26439518 | 0.05 |
ENSDART00000041787
|
trim35-28
|
tripartite motif containing 35-28 |
| chr13_+_31177934 | 0.05 |
ENSDART00000144568
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr6_+_6780873 | 0.05 |
ENSDART00000011865
|
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr2_+_12255568 | 0.05 |
ENSDART00000184164
ENSDART00000013454 |
prtfdc1
|
phosphoribosyl transferase domain containing 1 |
| chr5_-_60885935 | 0.04 |
ENSDART00000128350
|
rad51d
|
RAD51 paralog D |
| chr3_-_56924654 | 0.04 |
ENSDART00000157038
|
hid1a
|
HID1 domain containing a |
| chr7_-_2163361 | 0.04 |
ENSDART00000173654
|
si:cabz01007812.1
|
si:cabz01007812.1 |
| chr9_+_13986427 | 0.04 |
ENSDART00000147200
|
cd28
|
CD28 molecule |
| chr17_+_1627379 | 0.04 |
ENSDART00000184050
|
LO018432.1
|
|
| chr25_-_20049449 | 0.04 |
ENSDART00000104315
|
zgc:136858
|
zgc:136858 |
| chr9_+_41612642 | 0.04 |
ENSDART00000138473
|
spegb
|
SPEG complex locus b |
| chr16_+_26747766 | 0.04 |
ENSDART00000183257
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr5_+_54422941 | 0.04 |
ENSDART00000175157
|
traf2b
|
Tnf receptor-associated factor 2b |
| chr24_+_13635108 | 0.04 |
ENSDART00000183008
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr20_-_14054083 | 0.04 |
ENSDART00000009549
|
rhag
|
Rh associated glycoprotein |
| chr2_+_32796873 | 0.03 |
ENSDART00000077511
|
ccr9a
|
chemokine (C-C motif) receptor 9a |
| chr17_+_15433671 | 0.03 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr16_+_11724230 | 0.03 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr24_+_37722048 | 0.03 |
ENSDART00000191918
|
rab11fip3
|
RAB11 family interacting protein 3 (class II) |
| chr11_+_8129536 | 0.03 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr16_+_38820660 | 0.03 |
ENSDART00000182538
ENSDART00000189879 |
trhra
|
thyrotropin-releasing hormone receptor a |
| chr11_+_30057762 | 0.02 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr5_-_41860556 | 0.02 |
ENSDART00000140154
|
si:dkey-65b12.12
|
si:dkey-65b12.12 |
| chr1_+_23398405 | 0.02 |
ENSDART00000102646
|
rhoh
|
ras homolog family member H |
| chr21_+_5915041 | 0.02 |
ENSDART00000151370
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr20_+_15015557 | 0.02 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr7_-_29571615 | 0.02 |
ENSDART00000019140
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr22_-_5323482 | 0.02 |
ENSDART00000145785
|
s1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr16_+_46410520 | 0.02 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr21_-_28439596 | 0.02 |
ENSDART00000089980
ENSDART00000132844 |
rasgrp2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr8_-_43997538 | 0.02 |
ENSDART00000186449
|
rimbp2
|
RIMS binding protein 2 |
| chr17_+_15433518 | 0.02 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr22_+_21255860 | 0.02 |
ENSDART00000134893
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr3_+_30968883 | 0.01 |
ENSDART00000085736
|
prf1.9
|
perforin 1.9 |
| chr24_+_19518570 | 0.01 |
ENSDART00000056081
|
sulf1
|
sulfatase 1 |
| chr11_+_38280454 | 0.01 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr9_+_17984358 | 0.01 |
ENSDART00000192399
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr8_+_36803415 | 0.01 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr16_+_26439939 | 0.01 |
ENSDART00000143073
|
trim35-28
|
tripartite motif containing 35-28 |
| chr7_-_7810348 | 0.01 |
ENSDART00000171984
|
cxcl19
|
chemokine (C-X-C motif) ligand 19 |
| chr10_-_9086716 | 0.01 |
ENSDART00000140095
|
arl15b
|
ADP-ribosylation factor-like 15b |
| chr15_+_29662401 | 0.01 |
ENSDART00000135540
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr21_-_21096437 | 0.01 |
ENSDART00000186552
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr6_-_40651944 | 0.00 |
ENSDART00000187423
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr12_+_18543783 | 0.00 |
ENSDART00000148326
ENSDART00000134530 |
mlst8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr2_-_5505094 | 0.00 |
ENSDART00000145035
|
saga
|
S-antigen; retina and pineal gland (arrestin) a |
| chr9_-_44939104 | 0.00 |
ENSDART00000192903
|
vil1
|
villin 1 |
| chr11_+_30058139 | 0.00 |
ENSDART00000112254
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr25_-_27564205 | 0.00 |
ENSDART00000157319
|
hyal4
|
hyaluronoglucosaminidase 4 |
| chr1_+_11736420 | 0.00 |
ENSDART00000140329
|
chst12b
|
carbohydrate (chondroitin 4) sulfotransferase 12b |
| chr2_+_38804223 | 0.00 |
ENSDART00000147939
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of bcl11aa
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.3 | GO:1900181 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.1 | 0.4 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 0.6 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.2 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.1 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.2 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.9 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.2 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.0 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.4 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.6 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.3 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |