Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for barx2
Z-value: 1.42
Transcription factors associated with barx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
barx2
|
ENSDARG00000041098 | BARX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| barx2 | dr11_v1_chr18_+_47313899_47313899 | 0.65 | 3.4e-03 | Click! |
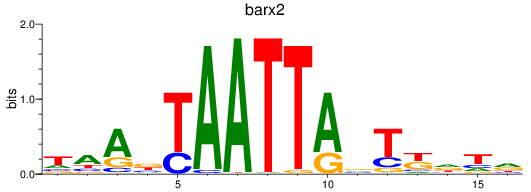
Activity profile of barx2 motif
Sorted Z-values of barx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_54209504 | 1.91 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr10_-_29236860 | 1.68 |
ENSDART00000111620
|
ccdc83
|
coiled-coil domain containing 83 |
| chr3_+_26145013 | 1.58 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr13_-_33683889 | 1.57 |
ENSDART00000136820
ENSDART00000065435 |
cst3
|
cystatin C (amyloid angiopathy and cerebral hemorrhage) |
| chr24_+_3478871 | 1.44 |
ENSDART00000111491
ENSDART00000134598 ENSDART00000142407 |
wdr37
|
WD repeat domain 37 |
| chr23_+_7710447 | 1.43 |
ENSDART00000168199
|
kif3b
|
kinesin family member 3B |
| chr4_+_17279966 | 1.42 |
ENSDART00000067005
ENSDART00000137487 |
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr17_-_39772999 | 1.38 |
ENSDART00000155727
|
pimr60
|
Pim proto-oncogene, serine/threonine kinase, related 60 |
| chr21_-_26490186 | 1.37 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr15_+_34988148 | 1.36 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr9_-_25055722 | 1.36 |
ENSDART00000137131
|
dnah7
|
dynein, axonemal, heavy chain 7 |
| chr23_-_36857964 | 1.33 |
ENSDART00000188822
ENSDART00000134061 ENSDART00000093061 |
hipk1a
|
homeodomain interacting protein kinase 1a |
| chr5_+_42386705 | 1.31 |
ENSDART00000143034
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr5_+_42407962 | 1.31 |
ENSDART00000188489
|
BX548073.11
|
|
| chr2_+_36004381 | 1.31 |
ENSDART00000098706
|
lamc2
|
laminin, gamma 2 |
| chr5_+_42400777 | 1.29 |
ENSDART00000183114
|
BX548073.8
|
|
| chr2_+_49799470 | 1.28 |
ENSDART00000146325
|
si:ch211-190k17.19
|
si:ch211-190k17.19 |
| chr3_+_60721342 | 1.25 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr5_+_42379517 | 1.24 |
ENSDART00000103325
|
pimr59
|
Pim proto-oncogene, serine/threonine kinase, related 59 |
| chr5_-_42661012 | 1.24 |
ENSDART00000158339
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr20_+_46040666 | 1.24 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr8_-_44586981 | 1.23 |
ENSDART00000026831
ENSDART00000113945 |
rsph14
|
radial spoke head 14 homolog |
| chr23_+_7710721 | 1.23 |
ENSDART00000186852
ENSDART00000161193 |
kif3b
|
kinesin family member 3B |
| chr15_-_20939579 | 1.20 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr17_-_39779906 | 1.20 |
ENSDART00000155181
|
pimr61
|
Pim proto-oncogene, serine/threonine kinase, related 61 |
| chr16_+_28994709 | 1.18 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr20_-_13640598 | 1.16 |
ENSDART00000128823
ENSDART00000103394 |
rsph3
|
radial spoke 3 homolog |
| chr9_-_24970018 | 1.15 |
ENSDART00000026924
|
dnah7
|
dynein, axonemal, heavy chain 7 |
| chr7_+_9981757 | 1.15 |
ENSDART00000113429
ENSDART00000173233 |
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr14_-_470505 | 1.13 |
ENSDART00000067147
|
ANKRD50
|
ankyrin repeat domain 50 |
| chr5_+_38040407 | 1.13 |
ENSDART00000139936
|
si:dkey-111e8.4
|
si:dkey-111e8.4 |
| chr5_-_54672763 | 1.10 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr5_+_42393896 | 1.09 |
ENSDART00000189550
|
BX548073.13
|
|
| chr24_+_35787629 | 1.08 |
ENSDART00000136721
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr13_+_7049823 | 1.07 |
ENSDART00000178997
ENSDART00000161443 |
rnaset2
|
ribonuclease T2 |
| chr13_+_24750078 | 1.06 |
ENSDART00000021053
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr13_-_38730267 | 1.06 |
ENSDART00000157524
|
lmbrd1
|
LMBR1 domain containing 1 |
| chr4_-_2219705 | 1.05 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr19_+_1688727 | 1.03 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr19_-_42045372 | 1.02 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr7_-_40110140 | 0.99 |
ENSDART00000173469
|
si:ch73-174h16.5
|
si:ch73-174h16.5 |
| chr15_-_18162647 | 0.99 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr14_-_1355544 | 0.99 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr8_-_53044300 | 0.99 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr25_-_21031007 | 0.98 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr20_+_33739059 | 0.98 |
ENSDART00000140361
|
ppil6
|
peptidylprolyl isomerase (cyclophilin)-like 6 |
| chr16_-_20932896 | 0.95 |
ENSDART00000180646
|
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr25_-_37180969 | 0.95 |
ENSDART00000152338
|
tdrd12
|
tudor domain containing 12 |
| chr15_-_25083200 | 0.94 |
ENSDART00000156053
|
pimr191
|
Pim proto-oncogene, serine/threonine kinase, related 191 |
| chr20_+_42881104 | 0.93 |
ENSDART00000131338
|
pimr110
|
Pim proto-oncogene, serine/threonine kinase, related 110 |
| chr4_+_9669717 | 0.92 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr14_-_48765262 | 0.91 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr9_-_36924388 | 0.89 |
ENSDART00000059756
|
ralba
|
v-ral simian leukemia viral oncogene homolog Ba (ras related) |
| chr22_+_508290 | 0.89 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr4_-_5019113 | 0.88 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr6_-_55254786 | 0.88 |
ENSDART00000113805
|
nfatc2b
|
nuclear factor of activated T cells 2b |
| chr21_+_25236297 | 0.88 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr7_+_19552381 | 0.87 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr11_-_27867024 | 0.86 |
ENSDART00000182136
ENSDART00000187587 |
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr3_-_12026741 | 0.86 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr21_-_36571804 | 0.85 |
ENSDART00000138129
|
wwc1
|
WW and C2 domain containing 1 |
| chr8_-_39822917 | 0.85 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr4_+_2482046 | 0.85 |
ENSDART00000103371
|
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr21_-_2814709 | 0.85 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr19_+_46158078 | 0.82 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr18_+_3579829 | 0.82 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr23_+_40460333 | 0.82 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr7_+_73397283 | 0.81 |
ENSDART00000174390
|
CABZ01081780.1
|
|
| chr24_+_28953089 | 0.80 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr22_-_37797695 | 0.79 |
ENSDART00000085931
ENSDART00000185443 |
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr2_-_1548330 | 0.78 |
ENSDART00000082155
ENSDART00000108481 ENSDART00000111272 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr23_-_33350990 | 0.78 |
ENSDART00000144831
|
si:ch211-226m16.2
|
si:ch211-226m16.2 |
| chr23_+_4253957 | 0.78 |
ENSDART00000008761
|
arl6ip5b
|
ADP-ribosylation factor-like 6 interacting protein 5b |
| chr7_-_40145097 | 0.78 |
ENSDART00000173634
|
wdr60
|
WD repeat domain 60 |
| chr19_-_3931917 | 0.76 |
ENSDART00000162532
|
map7d1b
|
MAP7 domain containing 1b |
| chr16_-_42894628 | 0.74 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr2_+_2169337 | 0.74 |
ENSDART00000179939
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr25_+_28825657 | 0.74 |
ENSDART00000153625
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr21_+_25802190 | 0.73 |
ENSDART00000128987
|
nf2b
|
neurofibromin 2b (merlin) |
| chr11_+_34921492 | 0.72 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr20_+_40457599 | 0.72 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr22_-_20812822 | 0.72 |
ENSDART00000193778
|
dot1l
|
DOT1-like histone H3K79 methyltransferase |
| chr19_-_3106447 | 0.72 |
ENSDART00000137987
|
si:ch211-80h18.1
|
si:ch211-80h18.1 |
| chr16_+_28728347 | 0.72 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr6_-_40744720 | 0.72 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr2_+_33751490 | 0.71 |
ENSDART00000145031
|
si:dkey-31m5.7
|
si:dkey-31m5.7 |
| chr16_-_35329803 | 0.71 |
ENSDART00000161729
ENSDART00000157700 ENSDART00000184584 ENSDART00000174713 ENSDART00000162518 |
ptprub
|
protein tyrosine phosphatase, receptor type, U, b |
| chr20_-_51355465 | 0.71 |
ENSDART00000151620
ENSDART00000151690 ENSDART00000110289 |
tcte1
|
t-complex-associated-testis-expressed 1 |
| chr22_-_37796998 | 0.70 |
ENSDART00000124742
ENSDART00000191232 |
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr9_-_47472998 | 0.69 |
ENSDART00000134480
|
tns1b
|
tensin 1b |
| chr16_+_40024883 | 0.69 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr5_+_63302660 | 0.69 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr22_+_30137374 | 0.68 |
ENSDART00000187808
|
add3a
|
adducin 3 (gamma) a |
| chr9_-_7652792 | 0.68 |
ENSDART00000137957
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr21_+_38745094 | 0.68 |
ENSDART00000113316
|
heatr6
|
HEAT repeat containing 6 |
| chr19_+_1673599 | 0.67 |
ENSDART00000163127
|
klhl7
|
kelch-like family member 7 |
| chr20_+_41756996 | 0.67 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr1_-_56080112 | 0.66 |
ENSDART00000075469
ENSDART00000161473 |
c3a.6
|
complement component c3a, duplicate 6 |
| chr16_-_28856112 | 0.66 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr18_-_48547564 | 0.65 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr13_-_48764180 | 0.65 |
ENSDART00000167157
|
si:ch1073-266p11.2
|
si:ch1073-266p11.2 |
| chr25_-_27722614 | 0.65 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr7_-_40144686 | 0.65 |
ENSDART00000109166
|
wdr60
|
WD repeat domain 60 |
| chr5_-_41494831 | 0.64 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr2_+_19522082 | 0.64 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr25_-_37186894 | 0.64 |
ENSDART00000191647
ENSDART00000182095 |
tdrd12
|
tudor domain containing 12 |
| chr11_+_31190155 | 0.63 |
ENSDART00000112098
|
stx10
|
syntaxin 10 |
| chr6_+_13117598 | 0.63 |
ENSDART00000104744
|
casp8l1
|
caspase 8, apoptosis-related cysteine peptidase, like 1 |
| chr17_+_6625717 | 0.63 |
ENSDART00000190753
ENSDART00000181298 |
BX005321.1
|
|
| chr8_+_7097740 | 0.63 |
ENSDART00000159670
|
abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr16_-_50203058 | 0.62 |
ENSDART00000154570
|
vsig10l
|
V-set and immunoglobulin domain containing 10 like |
| chr15_-_39955785 | 0.62 |
ENSDART00000154556
|
msh5
|
mutS homolog 5 |
| chr7_-_31922432 | 0.62 |
ENSDART00000188398
|
lin7c
|
lin-7 homolog C (C. elegans) |
| chr13_-_9450210 | 0.61 |
ENSDART00000133768
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr24_+_7336807 | 0.60 |
ENSDART00000137010
|
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr6_+_29305190 | 0.60 |
ENSDART00000078647
|
si:ch211-201h21.5
|
si:ch211-201h21.5 |
| chr1_+_27977297 | 0.60 |
ENSDART00000180692
ENSDART00000166819 |
sugt1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr7_+_49681040 | 0.60 |
ENSDART00000176372
ENSDART00000192172 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr6_-_29305132 | 0.60 |
ENSDART00000132456
|
bivm
|
basic, immunoglobulin-like variable motif containing |
| chr11_+_31121340 | 0.60 |
ENSDART00000185172
|
stx10
|
syntaxin 10 |
| chr5_-_11809710 | 0.60 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr5_+_63668735 | 0.60 |
ENSDART00000134261
ENSDART00000097330 |
dnm1b
|
dynamin 1b |
| chr13_+_26703922 | 0.59 |
ENSDART00000020946
|
fancl
|
Fanconi anemia, complementation group L |
| chr10_-_34046757 | 0.59 |
ENSDART00000099648
|
pimr149
|
Pim proto-oncogene, serine/threonine kinase, related 149 |
| chr13_+_35339182 | 0.59 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr16_+_54263921 | 0.57 |
ENSDART00000002856
|
drd2l
|
dopamine receptor D2 like |
| chr1_-_19502322 | 0.57 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr17_+_41992054 | 0.56 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr8_-_50888806 | 0.56 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr7_+_26534131 | 0.56 |
ENSDART00000173980
|
si:dkey-62k3.5
|
si:dkey-62k3.5 |
| chr15_-_33965440 | 0.56 |
ENSDART00000163841
|
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr2_-_1622641 | 0.55 |
ENSDART00000082143
|
prkacbb
|
protein kinase, cAMP-dependent, catalytic, beta b |
| chr16_-_34212912 | 0.55 |
ENSDART00000145017
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr2_+_33726862 | 0.55 |
ENSDART00000146745
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr16_-_21140097 | 0.55 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr1_-_46981134 | 0.54 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr4_-_9909371 | 0.54 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr9_+_35832294 | 0.54 |
ENSDART00000130549
ENSDART00000122169 |
si:dkey-67c22.2
|
si:dkey-67c22.2 |
| chr7_+_66884570 | 0.54 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr3_+_40284598 | 0.54 |
ENSDART00000009411
|
bud31
|
BUD31 homolog (S. cerevisiae) |
| chr21_-_32097908 | 0.53 |
ENSDART00000147387
|
si:ch211-160j14.3
|
si:ch211-160j14.3 |
| chr7_+_20471315 | 0.53 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr23_+_4709607 | 0.52 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr5_+_2815021 | 0.52 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr10_-_34123733 | 0.52 |
ENSDART00000157708
ENSDART00000143954 |
pimr150
|
Pim proto-oncogene, serine/threonine kinase, related 150 |
| chr3_+_17030665 | 0.52 |
ENSDART00000159849
ENSDART00000174491 ENSDART00000104519 ENSDART00000080854 |
stat3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr10_+_15967643 | 0.51 |
ENSDART00000136709
|
apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr16_-_44945224 | 0.51 |
ENSDART00000156921
|
ncam3
|
neural cell adhesion molecule 3 |
| chr17_+_12285285 | 0.51 |
ENSDART00000154336
|
pimr174
|
Pim proto-oncogene, serine/threonine kinase, related 174 |
| chr2_-_33645411 | 0.51 |
ENSDART00000114663
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr3_+_62290024 | 0.50 |
ENSDART00000185108
|
zgc:173575
|
zgc:173575 |
| chr7_+_1530024 | 0.50 |
ENSDART00000163082
|
tox4b
|
TOX high mobility group box family member 4 b |
| chr4_+_16715267 | 0.50 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr5_+_10084100 | 0.50 |
ENSDART00000109236
|
si:ch211-207k7.4
|
si:ch211-207k7.4 |
| chr12_+_18578597 | 0.50 |
ENSDART00000134944
|
grid2ipb
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b |
| chr13_-_9467944 | 0.49 |
ENSDART00000136582
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr6_+_25257728 | 0.49 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr7_+_24153070 | 0.49 |
ENSDART00000076735
|
lrp10
|
low density lipoprotein receptor-related protein 10 |
| chr14_-_7137808 | 0.49 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr15_-_40267485 | 0.48 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr7_+_66884291 | 0.48 |
ENSDART00000187499
|
sbf2
|
SET binding factor 2 |
| chr3_-_40955780 | 0.48 |
ENSDART00000130130
|
cyp3c3
|
cytochrome P450, family 3, subfamily c, polypeptide 3 |
| chr18_+_35128685 | 0.48 |
ENSDART00000151579
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr3_+_22443313 | 0.47 |
ENSDART00000156450
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr12_-_35885349 | 0.47 |
ENSDART00000085662
|
cep131
|
centrosomal protein 131 |
| chr13_+_646700 | 0.47 |
ENSDART00000006892
|
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr14_-_16082806 | 0.47 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr24_+_12983434 | 0.46 |
ENSDART00000145214
ENSDART00000146911 ENSDART00000066700 |
eloca
|
elongin C paralog a |
| chr2_-_42065069 | 0.46 |
ENSDART00000140188
|
cspp1b
|
centrosome and spindle pole associated protein 1b |
| chr11_-_2838699 | 0.46 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr13_-_33227411 | 0.46 |
ENSDART00000057386
|
golga5
|
golgin A5 |
| chr25_-_25058508 | 0.45 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr13_+_9213805 | 0.45 |
ENSDART00000131455
|
pimr158
|
Pim proto-oncogene, serine/threonine kinase, related 158 |
| chr25_+_5068442 | 0.45 |
ENSDART00000097522
|
parvg
|
parvin, gamma |
| chr2_-_10877765 | 0.45 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr22_-_38274995 | 0.45 |
ENSDART00000179786
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr8_-_41264502 | 0.45 |
ENSDART00000133124
|
rnf10
|
ring finger protein 10 |
| chr2_+_7295515 | 0.44 |
ENSDART00000152987
|
si:dkeyp-106c3.1
|
si:dkeyp-106c3.1 |
| chr20_-_36408836 | 0.44 |
ENSDART00000076419
|
lbr
|
lamin B receptor |
| chr14_+_35892802 | 0.44 |
ENSDART00000135079
|
zgc:63568
|
zgc:63568 |
| chr1_+_55293424 | 0.44 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr20_-_47188966 | 0.44 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr1_-_669717 | 0.44 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr17_-_33412868 | 0.43 |
ENSDART00000187521
|
BX323819.1
|
|
| chr22_+_18816662 | 0.43 |
ENSDART00000132476
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr1_+_54037077 | 0.43 |
ENSDART00000109386
|
triobpa
|
TRIO and F-actin binding protein a |
| chr16_+_25074029 | 0.43 |
ENSDART00000155465
|
si:dkeyp-84f3.9
|
si:dkeyp-84f3.9 |
| chr13_-_52089003 | 0.43 |
ENSDART00000187600
|
tmem254
|
transmembrane protein 254 |
| chr8_-_20230802 | 0.42 |
ENSDART00000063400
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr7_-_25895189 | 0.42 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr16_+_29492937 | 0.42 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr14_+_46313396 | 0.42 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr6_+_9427641 | 0.42 |
ENSDART00000022620
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr19_+_43297546 | 0.41 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr20_-_53078607 | 0.41 |
ENSDART00000163494
ENSDART00000191730 |
CABZ01066813.1
|
|
| chr1_-_42289704 | 0.41 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr11_+_30057762 | 0.41 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr25_-_10503043 | 0.41 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr15_+_21262917 | 0.40 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr2_-_50198328 | 0.40 |
ENSDART00000074511
ENSDART00000137704 |
ahrrb
|
aryl-hydrocarbon receptor repressor b |
Network of associatons between targets according to the STRING database.
First level regulatory network of barx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0031446 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.4 | 1.4 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.3 | 1.7 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.3 | 2.7 | GO:0043584 | nose development(GO:0043584) |
| 0.2 | 1.2 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 0.7 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.2 | 0.5 | GO:0044321 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.2 | 1.0 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.1 | 0.9 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 0.6 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.8 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 1.0 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.7 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.9 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 0.5 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 0.3 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 0.8 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.3 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 1.1 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.5 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.4 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 2.5 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.1 | 0.3 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.1 | 0.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.3 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 1.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 1.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.5 | GO:0090317 | intraciliary transport involved in cilium morphogenesis(GO:0035735) negative regulation of intracellular protein transport(GO:0090317) |
| 0.1 | 0.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.4 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.1 | 1.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.7 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 0.9 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.1 | 0.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.3 | GO:0031652 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 0.3 | GO:0008591 | neural plate morphogenesis(GO:0001839) regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.1 | 0.6 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.6 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.4 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.2 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.1 | 0.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.7 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.9 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 1.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 0.7 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.5 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.7 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 1.4 | GO:0030814 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.3 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.0 | 0.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 13.2 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.3 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.3 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 1.1 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 1.1 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 2.6 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.7 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.6 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.1 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 1.3 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.3 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.3 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.7 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.3 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.3 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.5 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 1.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.4 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.4 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.1 | GO:1903400 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.3 | GO:0098781 | ncRNA transcription(GO:0098781) |
| 0.0 | 0.8 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 0.8 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.3 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0031673 | H zone(GO:0031673) |
| 0.4 | 1.1 | GO:0043202 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.2 | 1.0 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 0.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.4 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.6 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.3 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 1.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 2.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 2.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.4 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 1.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.5 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.8 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.4 | 1.4 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.4 | 1.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.3 | 0.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 0.8 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.2 | 1.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 0.7 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.2 | 0.5 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.2 | 0.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.3 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 1.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 2.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.5 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.3 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.7 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 2.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.6 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 14.4 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.3 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.2 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.0 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.3 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 1.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 4.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 2.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 1.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |