Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
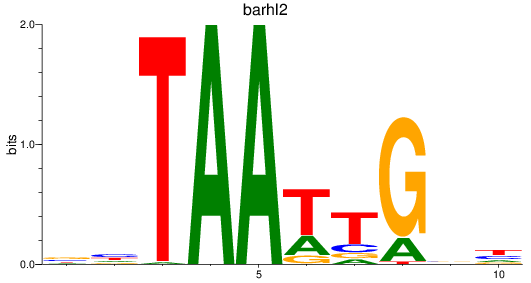
Results for barhl2
Z-value: 0.39
Transcription factors associated with barhl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
barhl2
|
ENSDARG00000104361 | BarH-like homeobox 2 |
|
barhl2
|
ENSDARG00000114681 | BarH-like homeobox 2 |
|
barhl2
|
ENSDARG00000115723 | BarH-like homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| barhl2 | dr11_v1_chr6_+_24817852_24817852 | -0.83 | 2.2e-05 | Click! |
Activity profile of barhl2 motif
Sorted Z-values of barhl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_17115256 | 1.67 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr6_-_8362419 | 1.59 |
ENSDART00000142752
ENSDART00000135810 |
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr21_+_25777425 | 1.28 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr1_-_33645967 | 1.19 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr8_+_45334255 | 0.98 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr21_-_19919020 | 0.89 |
ENSDART00000147396
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr7_+_28724919 | 0.88 |
ENSDART00000011324
|
ccdc102a
|
coiled-coil domain containing 102A |
| chr5_+_29794058 | 0.87 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr11_-_6452444 | 0.85 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr24_-_10006158 | 0.85 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr3_+_43102010 | 0.80 |
ENSDART00000162096
|
micall2a
|
mical-like 2a |
| chr19_-_874888 | 0.77 |
ENSDART00000007206
|
eomesa
|
eomesodermin homolog a |
| chr1_-_55248496 | 0.71 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr6_+_21001264 | 0.71 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr17_+_16046314 | 0.70 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr20_-_49889111 | 0.69 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr16_+_47207691 | 0.68 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr12_-_11349899 | 0.68 |
ENSDART00000079645
|
zgc:174164
|
zgc:174164 |
| chr17_+_16046132 | 0.68 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr17_+_43867889 | 0.68 |
ENSDART00000132673
ENSDART00000167214 |
zgc:66313
|
zgc:66313 |
| chr9_-_35633827 | 0.68 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr4_-_4259079 | 0.68 |
ENSDART00000135352
ENSDART00000026559 |
cd9b
|
CD9 molecule b |
| chr11_-_44801968 | 0.68 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr20_+_27087539 | 0.67 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr7_-_26270014 | 0.67 |
ENSDART00000079347
|
serpine1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr5_-_29512538 | 0.67 |
ENSDART00000098364
|
ehmt1a
|
euchromatic histone-lysine N-methyltransferase 1a |
| chr17_+_21887823 | 0.66 |
ENSDART00000131929
ENSDART00000165192 |
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr22_-_22147375 | 0.65 |
ENSDART00000149304
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr7_+_34487833 | 0.64 |
ENSDART00000173854
|
cln6a
|
CLN6, transmembrane ER protein a |
| chr5_-_72390259 | 0.62 |
ENSDART00000172302
|
wbp1
|
WW domain binding protein 1 |
| chr3_+_43373867 | 0.62 |
ENSDART00000159455
ENSDART00000172425 |
zfand2a
|
zinc finger, AN1-type domain 2A |
| chr9_+_500052 | 0.62 |
ENSDART00000166707
|
CU984600.1
|
|
| chr15_+_22394074 | 0.62 |
ENSDART00000109931
|
oafa
|
OAF homolog a (Drosophila) |
| chr1_+_513986 | 0.60 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr11_+_11303458 | 0.60 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr11_+_18130300 | 0.59 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr17_-_30635298 | 0.58 |
ENSDART00000155478
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr3_-_54607166 | 0.58 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr6_+_19948043 | 0.57 |
ENSDART00000182636
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr3_-_40232615 | 0.57 |
ENSDART00000155969
|
flii
|
flightless I actin binding protein |
| chr10_+_15024772 | 0.56 |
ENSDART00000135667
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr14_+_24845941 | 0.56 |
ENSDART00000187513
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr17_+_25849332 | 0.56 |
ENSDART00000191994
|
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr5_+_6670945 | 0.56 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr22_-_20695237 | 0.55 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr19_-_25119443 | 0.55 |
ENSDART00000148953
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr19_-_25149034 | 0.55 |
ENSDART00000148432
ENSDART00000175266 |
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr20_+_14789305 | 0.55 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr22_-_14272699 | 0.53 |
ENSDART00000190121
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr1_+_2301961 | 0.52 |
ENSDART00000108919
ENSDART00000143361 ENSDART00000142944 |
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr4_-_20081621 | 0.51 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr15_+_24795473 | 0.51 |
ENSDART00000139689
ENSDART00000141033 ENSDART00000100746 ENSDART00000135677 |
gosr1
|
golgi SNAP receptor complex member 1 |
| chr23_+_2666944 | 0.51 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr12_+_35203091 | 0.51 |
ENSDART00000153022
|
ndst2b
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2b |
| chr17_+_16429826 | 0.50 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr9_-_6991650 | 0.50 |
ENSDART00000081718
|
slc9a2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr18_+_8917766 | 0.50 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr11_+_18157260 | 0.50 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr17_-_14774117 | 0.50 |
ENSDART00000080401
|
si:ch211-266o15.1
|
si:ch211-266o15.1 |
| chr11_-_35171768 | 0.50 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr12_+_14149686 | 0.48 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr14_+_8940326 | 0.48 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr3_+_18807006 | 0.47 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr1_+_21937201 | 0.47 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr24_+_12835935 | 0.47 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr24_-_36238054 | 0.47 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr21_+_34119759 | 0.47 |
ENSDART00000024750
ENSDART00000128242 |
hmgb3b
|
high mobility group box 3b |
| chr7_+_24528430 | 0.47 |
ENSDART00000133022
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr1_-_18811517 | 0.46 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr11_+_45286911 | 0.46 |
ENSDART00000181763
|
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr17_-_6613458 | 0.46 |
ENSDART00000175024
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr23_+_2917392 | 0.46 |
ENSDART00000150019
|
DHX35
|
zgc:158828 |
| chr5_-_54554583 | 0.46 |
ENSDART00000158865
ENSDART00000158069 |
ssna1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr8_-_19467011 | 0.46 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr10_-_13343831 | 0.45 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr15_+_11644866 | 0.45 |
ENSDART00000188716
|
slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr15_-_30857350 | 0.44 |
ENSDART00000138988
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr5_-_42180205 | 0.44 |
ENSDART00000145247
|
fam222ba
|
family with sequence similarity 222, member Ba |
| chr7_+_38809241 | 0.43 |
ENSDART00000190979
|
harbi1
|
harbinger transposase derived 1 |
| chr16_-_7793457 | 0.43 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr6_-_39631164 | 0.43 |
ENSDART00000104042
ENSDART00000136076 |
atf7b
|
activating transcription factor 7b |
| chr23_+_38171186 | 0.43 |
ENSDART00000148188
|
zgc:112994
|
zgc:112994 |
| chr9_-_32300783 | 0.43 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr17_+_1544903 | 0.42 |
ENSDART00000156244
ENSDART00000112183 |
cep170b
|
centrosomal protein 170B |
| chr5_-_69620722 | 0.42 |
ENSDART00000097248
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr14_+_21820034 | 0.42 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr22_-_10541372 | 0.42 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr24_-_2450597 | 0.41 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr1_-_53407448 | 0.41 |
ENSDART00000160033
ENSDART00000172322 |
elmod2
|
ELMO/CED-12 domain containing 2 |
| chr20_-_23426339 | 0.41 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr22_+_16293071 | 0.41 |
ENSDART00000170960
|
dbt
|
dihydrolipoamide branched chain transacylase E2 |
| chr2_-_32387441 | 0.41 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr3_-_26244256 | 0.41 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr15_-_34408777 | 0.40 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr24_-_40901410 | 0.40 |
ENSDART00000170688
|
wdr48a
|
WD repeat domain 48a |
| chr16_+_38940758 | 0.40 |
ENSDART00000102482
ENSDART00000136215 |
eny2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr24_-_25166720 | 0.40 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr10_-_31805923 | 0.40 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr16_+_40340222 | 0.40 |
ENSDART00000190631
|
mettl6
|
methyltransferase like 6 |
| chr5_-_54395488 | 0.40 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr17_+_46387086 | 0.39 |
ENSDART00000157079
|
si:dkey-206p8.1
|
si:dkey-206p8.1 |
| chr13_-_31687925 | 0.39 |
ENSDART00000085989
|
trmt5
|
TRM5 tRNA methyltransferase 5 homolog (S. cerevisiae) |
| chr15_-_6966221 | 0.39 |
ENSDART00000165487
ENSDART00000027657 |
mrps22
|
mitochondrial ribosomal protein S22 |
| chr2_-_57076687 | 0.39 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr22_+_14117078 | 0.39 |
ENSDART00000013575
|
bzw1a
|
basic leucine zipper and W2 domains 1a |
| chr3_+_26244353 | 0.38 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr9_-_32300611 | 0.38 |
ENSDART00000127938
|
hspd1
|
heat shock 60 protein 1 |
| chr23_-_36303216 | 0.38 |
ENSDART00000188720
|
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr15_-_25269028 | 0.38 |
ENSDART00000078230
ENSDART00000193872 |
mettl16
|
methyltransferase like 16 |
| chr24_-_34680956 | 0.37 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr10_-_15854743 | 0.37 |
ENSDART00000092343
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr19_+_29808471 | 0.37 |
ENSDART00000186428
|
hdac1
|
histone deacetylase 1 |
| chr7_-_64971839 | 0.37 |
ENSDART00000164682
|
sinhcafl
|
SIN3-HDAC complex associated factor, like |
| chr9_-_14273652 | 0.37 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr20_-_48700370 | 0.37 |
ENSDART00000185195
ENSDART00000184898 |
pax1b
|
paired box 1b |
| chr11_+_45287541 | 0.37 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr16_+_54209504 | 0.36 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr18_+_3579829 | 0.36 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr7_+_61764040 | 0.36 |
ENSDART00000056745
|
acox3
|
acyl-CoA oxidase 3, pristanoyl |
| chr23_+_32029304 | 0.36 |
ENSDART00000185217
|
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr2_-_55779927 | 0.36 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr17_+_50701748 | 0.35 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr9_-_30385059 | 0.35 |
ENSDART00000060134
|
piga
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr19_+_40069524 | 0.34 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr16_+_40340523 | 0.34 |
ENSDART00000102571
|
mettl6
|
methyltransferase like 6 |
| chr7_-_40959867 | 0.33 |
ENSDART00000174009
|
rbm33a
|
RNA binding motif protein 33a |
| chr5_+_29652513 | 0.33 |
ENSDART00000035400
|
tsc1a
|
TSC complex subunit 1a |
| chr11_+_19370717 | 0.33 |
ENSDART00000165906
|
prickle2b
|
prickle homolog 2b |
| chr22_+_34701848 | 0.33 |
ENSDART00000082066
|
atpv0e2
|
ATPase H+ transporting V0 subunit e2 |
| chr9_+_29431763 | 0.33 |
ENSDART00000186095
ENSDART00000182640 |
uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr14_+_50937757 | 0.32 |
ENSDART00000163865
|
rnf44
|
ring finger protein 44 |
| chr12_+_33395748 | 0.32 |
ENSDART00000129458
|
fasn
|
fatty acid synthase |
| chr3_+_22035863 | 0.32 |
ENSDART00000177169
|
cdc27
|
cell division cycle 27 |
| chr2_+_6253246 | 0.32 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr10_-_14929630 | 0.31 |
ENSDART00000121892
ENSDART00000044756 ENSDART00000128579 ENSDART00000147653 |
smad2
|
SMAD family member 2 |
| chr14_+_26247319 | 0.31 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr20_+_18703454 | 0.31 |
ENSDART00000152342
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr15_-_26931541 | 0.31 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr11_+_19080400 | 0.31 |
ENSDART00000044423
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr20_-_34801181 | 0.31 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr5_-_35252761 | 0.31 |
ENSDART00000051278
|
tnpo1
|
transportin 1 |
| chr24_+_39227519 | 0.30 |
ENSDART00000184611
ENSDART00000193494 ENSDART00000190728 ENSDART00000168705 |
MPRIP
|
si:ch73-103b11.2 |
| chr19_-_34873566 | 0.30 |
ENSDART00000016057
|
ctnnal1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr19_+_15443540 | 0.30 |
ENSDART00000193355
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr14_-_46113321 | 0.30 |
ENSDART00000169040
ENSDART00000161475 ENSDART00000124925 |
si:ch211-235e9.8
|
si:ch211-235e9.8 |
| chr5_+_53580846 | 0.30 |
ENSDART00000184967
ENSDART00000161751 |
taf6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr13_-_533243 | 0.30 |
ENSDART00000038315
|
nvl
|
nuclear VCP-like |
| chr19_+_20793237 | 0.30 |
ENSDART00000014774
|
txnl4a
|
thioredoxin-like 4A |
| chr13_-_4018888 | 0.30 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr6_-_51771634 | 0.30 |
ENSDART00000073847
|
blcap
|
bladder cancer associated protein |
| chr7_-_51368681 | 0.30 |
ENSDART00000146385
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr23_+_24973773 | 0.30 |
ENSDART00000047020
|
casp9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr25_-_32869794 | 0.29 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr16_-_35427060 | 0.29 |
ENSDART00000172294
|
ctps1b
|
CTP synthase 1b |
| chr8_-_38201415 | 0.29 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr23_-_24247672 | 0.29 |
ENSDART00000141552
|
zbtb17
|
zinc finger and BTB domain containing 17 |
| chr6_-_10037207 | 0.29 |
ENSDART00000179701
|
nudt15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr11_+_25693395 | 0.29 |
ENSDART00000110224
|
mon1bb
|
MON1 secretory trafficking family member Bb |
| chr15_+_24676905 | 0.29 |
ENSDART00000078014
ENSDART00000143137 |
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr4_-_20108833 | 0.29 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr19_-_19871211 | 0.29 |
ENSDART00000170980
|
evx1
|
even-skipped homeobox 1 |
| chr15_+_1534644 | 0.29 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr11_+_1522136 | 0.29 |
ENSDART00000002318
|
srsf6b
|
serine/arginine-rich splicing factor 6b |
| chr18_-_34549721 | 0.28 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr18_-_18942098 | 0.28 |
ENSDART00000100458
|
si:dkey-73n10.1
|
si:dkey-73n10.1 |
| chr10_-_44560165 | 0.27 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr20_+_34671386 | 0.27 |
ENSDART00000152836
ENSDART00000138226 |
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr22_+_786556 | 0.27 |
ENSDART00000125347
|
cry1bb
|
cryptochrome circadian clock 1bb |
| chr10_+_17371356 | 0.27 |
ENSDART00000122663
|
sppl3
|
signal peptide peptidase 3 |
| chr16_-_25680666 | 0.27 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr8_-_50888806 | 0.27 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr19_-_8768564 | 0.27 |
ENSDART00000170416
|
si:ch73-350k19.1
|
si:ch73-350k19.1 |
| chr22_+_883678 | 0.27 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr6_-_42336987 | 0.26 |
ENSDART00000128777
ENSDART00000075601 |
fancd2
|
Fanconi anemia, complementation group D2 |
| chr7_+_24153070 | 0.26 |
ENSDART00000076735
|
lrp10
|
low density lipoprotein receptor-related protein 10 |
| chr11_-_25461336 | 0.26 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr21_+_43702016 | 0.26 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr23_-_15916316 | 0.26 |
ENSDART00000134096
ENSDART00000042469 ENSDART00000146605 |
mrgbp
|
MRG/MORF4L binding protein |
| chr16_-_13680692 | 0.26 |
ENSDART00000047452
|
ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr19_+_29808699 | 0.25 |
ENSDART00000051799
ENSDART00000164205 |
hdac1
|
histone deacetylase 1 |
| chr2_-_54039293 | 0.25 |
ENSDART00000166013
|
abhd8a
|
abhydrolase domain containing 8a |
| chr22_+_34784075 | 0.25 |
ENSDART00000167538
|
lcor
|
ligand dependent nuclear receptor corepressor |
| chr6_-_3982783 | 0.25 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr6_+_40922572 | 0.24 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr13_+_6188759 | 0.24 |
ENSDART00000161062
|
ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr3_+_32791786 | 0.24 |
ENSDART00000180174
|
tbc1d10b
|
TBC1 domain family, member 10b |
| chr20_+_18703108 | 0.24 |
ENSDART00000181188
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr13_+_1100197 | 0.24 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr20_+_18703695 | 0.24 |
ENSDART00000137766
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr16_-_24642814 | 0.24 |
ENSDART00000153987
ENSDART00000154319 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr20_-_54924593 | 0.24 |
ENSDART00000151522
|
si:dkey-15f23.1
|
si:dkey-15f23.1 |
| chr8_-_10949847 | 0.24 |
ENSDART00000123209
|
pqlc2
|
PQ loop repeat containing 2 |
| chr2_+_16652238 | 0.24 |
ENSDART00000091351
|
gk5
|
glycerol kinase 5 (putative) |
| chr10_-_33297864 | 0.23 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr21_+_27302752 | 0.23 |
ENSDART00000012855
|
sart1
|
SART1, U4/U6.U5 tri-snRNP-associated protein 1 |
| chr25_+_17590095 | 0.23 |
ENSDART00000009767
|
vac14
|
vac14 homolog (S. cerevisiae) |
| chr2_-_38206034 | 0.23 |
ENSDART00000144518
ENSDART00000137395 |
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr10_+_43039947 | 0.23 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr3_-_32873641 | 0.23 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr11_-_40457325 | 0.22 |
ENSDART00000128442
|
tnfrsf1b
|
tumor necrosis factor receptor superfamily, member 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of barhl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 | 0.9 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 0.8 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.2 | 0.8 | GO:0045041 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 0.7 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 1.7 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 1.6 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 0.4 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.4 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.5 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.5 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.1 | 0.3 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.4 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.5 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.6 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 0.8 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.3 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.1 | 0.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.6 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.1 | 0.4 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.3 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.1 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.4 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.7 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.2 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.1 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.1 | 0.2 | GO:0071042 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.1 | 0.5 | GO:0006971 | hypotonic response(GO:0006971) hypotonic salinity response(GO:0042539) |
| 0.1 | 0.3 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.3 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.2 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.9 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.0 | 0.2 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.4 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.6 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.3 | GO:0006203 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 0.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.0 | 0.2 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 | 0.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.7 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.3 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.4 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.2 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.3 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.4 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.3 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.4 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) |
| 0.0 | 0.8 | GO:0097205 | renal filtration(GO:0097205) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.0 | GO:0070169 | positive regulation of bone mineralization(GO:0030501) positive regulation of ossification(GO:0045778) positive regulation of biomineral tissue development(GO:0070169) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.3 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.3 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.2 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.6 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.6 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.2 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.1 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.9 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.2 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.6 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.5 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.0 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.4 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.4 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.3 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 0.3 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 0.3 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 1.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.3 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 1.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.6 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0033065 | Rad51C-XRCC3 complex(GO:0033065) |
| 0.0 | 1.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.2 | GO:0035032 | extrinsic component of vacuolar membrane(GO:0000306) phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 3.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 1.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 1.0 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.2 | 0.8 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.4 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.4 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.1 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.5 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.3 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) |
| 0.1 | 0.2 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.4 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 1.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.0 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.3 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.2 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.1 | 0.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.8 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.5 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.5 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.4 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 1.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.0 | GO:0061650 | ubiquitin-like protein conjugating enzyme activity(GO:0061650) UFM1 transferase activity(GO:0071568) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 3.2 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 1.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |