Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
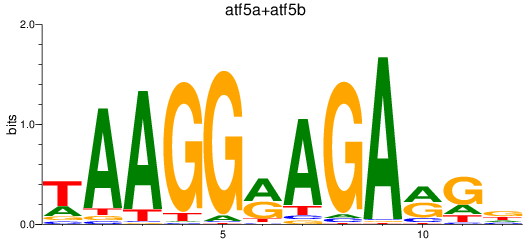
Results for atf5a+atf5b
Z-value: 0.66
Transcription factors associated with atf5a+atf5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf5a
|
ENSDARG00000068096 | activating transcription factor 5a |
|
atf5b
|
ENSDARG00000077785 | activating transcription factor 5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf5a | dr11_v1_chr5_-_30418636_30418636 | -0.85 | 9.9e-06 | Click! |
| atf5b | dr11_v1_chr15_-_17869115_17869115 | 0.75 | 3.6e-04 | Click! |
Activity profile of atf5a+atf5b motif
Sorted Z-values of atf5a+atf5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_42027969 | 2.82 |
ENSDART00000147563
|
kiz
|
kizuna centrosomal protein |
| chr5_+_42467867 | 1.87 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr10_-_34889053 | 1.62 |
ENSDART00000136966
|
ccdc169
|
coiled-coil domain containing 169 |
| chr16_+_10918252 | 1.51 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr7_-_28147838 | 1.42 |
ENSDART00000158921
|
lmo1
|
LIM domain only 1 |
| chr3_-_16227683 | 1.20 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr1_+_54626491 | 1.19 |
ENSDART00000136063
|
si:ch211-202h22.9
|
si:ch211-202h22.9 |
| chr5_+_30384554 | 1.18 |
ENSDART00000135483
|
zgc:158412
|
zgc:158412 |
| chr5_-_1963498 | 1.09 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr6_+_49053319 | 1.07 |
ENSDART00000124524
|
sycp1
|
synaptonemal complex protein 1 |
| chr12_+_35046704 | 1.06 |
ENSDART00000105523
ENSDART00000149946 |
timm23a
|
translocase of inner mitochondrial membrane 23 homolog a (yeast) |
| chr15_+_36977208 | 1.06 |
ENSDART00000183625
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr19_-_3931917 | 1.06 |
ENSDART00000162532
|
map7d1b
|
MAP7 domain containing 1b |
| chr24_+_9372292 | 1.06 |
ENSDART00000082422
ENSDART00000191127 ENSDART00000180510 |
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr11_+_5926850 | 1.05 |
ENSDART00000104364
|
rps15
|
ribosomal protein S15 |
| chr11_-_29657947 | 1.05 |
ENSDART00000125753
|
rpl22
|
ribosomal protein L22 |
| chr22_+_15624371 | 1.03 |
ENSDART00000124868
|
lpl
|
lipoprotein lipase |
| chr3_+_26081343 | 0.98 |
ENSDART00000134647
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr16_-_24518027 | 0.97 |
ENSDART00000134120
ENSDART00000143761 |
cadm4
|
cell adhesion molecule 4 |
| chr15_+_22267847 | 0.91 |
ENSDART00000110665
|
spa17
|
sperm autoantigenic protein 17 |
| chr1_-_40123943 | 0.87 |
ENSDART00000146917
|
si:ch211-113e8.10
|
si:ch211-113e8.10 |
| chr23_-_35069805 | 0.84 |
ENSDART00000087219
|
BX294434.1
|
|
| chr25_-_32751982 | 0.79 |
ENSDART00000012862
|
isl2a
|
ISL LIM homeobox 2a |
| chr20_-_26420939 | 0.78 |
ENSDART00000110883
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr3_-_16227490 | 0.76 |
ENSDART00000057159
ENSDART00000130611 ENSDART00000012835 |
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr18_+_19456648 | 0.74 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr13_-_23007813 | 0.73 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr4_-_64284924 | 0.72 |
ENSDART00000166733
|
CT956002.2
|
|
| chr16_+_29303971 | 0.72 |
ENSDART00000087149
|
hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr20_-_26421112 | 0.71 |
ENSDART00000183767
ENSDART00000182330 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr11_+_16152316 | 0.71 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr18_-_19456269 | 0.70 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr1_-_16665044 | 0.70 |
ENSDART00000040434
|
asah1b
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1b |
| chr19_+_3826782 | 0.70 |
ENSDART00000169222
|
oscp1a
|
organic solute carrier partner 1a |
| chr16_-_9802998 | 0.70 |
ENSDART00000154217
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr7_-_20758825 | 0.68 |
ENSDART00000156717
ENSDART00000182629 ENSDART00000179801 |
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr20_+_51478939 | 0.68 |
ENSDART00000149758
|
tlr5a
|
toll-like receptor 5a |
| chr11_-_25324534 | 0.68 |
ENSDART00000158598
|
si:ch211-232b12.5
|
si:ch211-232b12.5 |
| chr23_-_31506854 | 0.67 |
ENSDART00000131352
ENSDART00000138625 ENSDART00000133002 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr1_-_37377509 | 0.67 |
ENSDART00000113542
|
tnip2
|
TNFAIP3 interacting protein 2 |
| chr11_+_26476153 | 0.66 |
ENSDART00000103507
|
unm_sa1614
|
un-named sa1614 |
| chr18_-_23875370 | 0.65 |
ENSDART00000130163
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr19_-_35596207 | 0.63 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr18_+_28988373 | 0.61 |
ENSDART00000018685
|
syt9a
|
synaptotagmin IXa |
| chr16_+_10346277 | 0.60 |
ENSDART00000081092
|
si:dkeyp-77h1.4
|
si:dkeyp-77h1.4 |
| chr11_+_25472758 | 0.60 |
ENSDART00000011178
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr5_-_19052184 | 0.59 |
ENSDART00000133330
|
fam214b
|
family with sequence similarity 214, member B |
| chr12_+_27024676 | 0.59 |
ENSDART00000153104
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr22_+_3045495 | 0.58 |
ENSDART00000164061
|
LO017843.1
|
|
| chr14_-_25949951 | 0.57 |
ENSDART00000141304
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr4_-_12104421 | 0.55 |
ENSDART00000139561
|
mrps33
|
mitochondrial ribosomal protein S33 |
| chr3_+_37707432 | 0.53 |
ENSDART00000151236
|
map3k14a
|
mitogen-activated protein kinase kinase kinase 14a |
| chr21_-_22115136 | 0.53 |
ENSDART00000134715
ENSDART00000089246 ENSDART00000139789 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr3_-_30123113 | 0.52 |
ENSDART00000153562
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr18_-_29977431 | 0.51 |
ENSDART00000135357
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr12_+_8373525 | 0.51 |
ENSDART00000152180
|
arid5b
|
AT-rich interaction domain 5B |
| chr9_-_32837860 | 0.50 |
ENSDART00000142227
|
mxe
|
myxovirus (influenza virus) resistance E |
| chr5_-_67911111 | 0.49 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr20_+_51479263 | 0.49 |
ENSDART00000148798
|
tlr5a
|
toll-like receptor 5a |
| chr11_-_16152400 | 0.49 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr13_+_28854438 | 0.48 |
ENSDART00000193407
ENSDART00000189554 |
CU639469.1
|
|
| chr10_-_43568239 | 0.47 |
ENSDART00000131731
ENSDART00000097433 ENSDART00000131309 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr20_+_6630540 | 0.47 |
ENSDART00000138361
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr20_+_30797329 | 0.46 |
ENSDART00000145066
|
nhsl1b
|
NHS-like 1b |
| chr21_-_40348790 | 0.45 |
ENSDART00000178123
|
CR847523.1
|
|
| chr8_-_14067517 | 0.45 |
ENSDART00000140948
|
dedd
|
death effector domain containing |
| chr10_-_15672862 | 0.44 |
ENSDART00000109231
|
mamdc2b
|
MAM domain containing 2b |
| chr18_+_17786548 | 0.44 |
ENSDART00000189493
ENSDART00000146133 |
ZNF423
|
si:ch211-216l23.1 |
| chr24_-_1341543 | 0.44 |
ENSDART00000169341
|
nrp1a
|
neuropilin 1a |
| chr9_+_32930622 | 0.43 |
ENSDART00000100928
|
cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr5_-_1047222 | 0.43 |
ENSDART00000181112
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr10_+_17714866 | 0.43 |
ENSDART00000039969
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr4_-_72609735 | 0.41 |
ENSDART00000174299
ENSDART00000159227 |
si:cabz01054394.6
|
si:cabz01054394.6 |
| chr5_-_48307804 | 0.40 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr21_-_38031038 | 0.40 |
ENSDART00000179483
ENSDART00000076238 |
rbm41
|
RNA binding motif protein 41 |
| chr7_-_52153105 | 0.40 |
ENSDART00000174378
|
CT737190.1
|
|
| chr25_-_13320986 | 0.39 |
ENSDART00000169238
|
si:ch211-194m7.8
|
si:ch211-194m7.8 |
| chr12_-_31995840 | 0.39 |
ENSDART00000112881
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr11_+_24046179 | 0.38 |
ENSDART00000006703
|
maf1
|
MAF1 homolog, negative regulator of RNA polymerase III |
| chr13_+_30228077 | 0.38 |
ENSDART00000100813
ENSDART00000147729 ENSDART00000133404 |
rps24
|
ribosomal protein S24 |
| chr3_+_23703704 | 0.38 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr11_-_30630628 | 0.37 |
ENSDART00000103265
|
zgc:158773
|
zgc:158773 |
| chr16_+_42018367 | 0.37 |
ENSDART00000058613
|
fli1b
|
Fli-1 proto-oncogene, ETS transcription factor b |
| chr7_+_60359347 | 0.37 |
ENSDART00000145201
ENSDART00000039827 |
ppp1r14bb
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Bb |
| chr11_-_16152105 | 0.37 |
ENSDART00000081062
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr12_+_47663419 | 0.36 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr5_-_33287691 | 0.36 |
ENSDART00000004238
|
rpl7a
|
ribosomal protein L7a |
| chr16_-_13004166 | 0.36 |
ENSDART00000133735
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr25_-_16832705 | 0.35 |
ENSDART00000171336
ENSDART00000189396 |
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr5_-_1047504 | 0.34 |
ENSDART00000159346
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr7_-_38087865 | 0.34 |
ENSDART00000052366
|
cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr5_-_26187097 | 0.34 |
ENSDART00000137027
|
ccdc125
|
coiled-coil domain containing 125 |
| chr18_-_50152689 | 0.33 |
ENSDART00000006078
|
loxl1
|
lysyl oxidase-like 1 |
| chr24_-_23671383 | 0.33 |
ENSDART00000144193
|
hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr14_-_16772452 | 0.32 |
ENSDART00000137493
|
mrnip
|
MRN complex interacting protein |
| chr4_-_16334362 | 0.32 |
ENSDART00000101461
|
epyc
|
epiphycan |
| chr8_-_7474997 | 0.32 |
ENSDART00000146555
|
gata1b
|
GATA binding protein 1b |
| chr4_-_965267 | 0.32 |
ENSDART00000093289
|
atp23
|
ATP23 metallopeptidase and ATP synthase assembly factor homolog |
| chr15_+_15390882 | 0.31 |
ENSDART00000062024
|
ca4b
|
carbonic anhydrase IV b |
| chr16_+_42018041 | 0.31 |
ENSDART00000134010
ENSDART00000102789 |
fli1b
|
Fli-1 proto-oncogene, ETS transcription factor b |
| chr7_+_31879649 | 0.31 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr6_-_39765546 | 0.31 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr22_+_36656680 | 0.31 |
ENSDART00000134031
ENSDART00000056169 |
b3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr4_+_15983541 | 0.30 |
ENSDART00000043660
|
copg2
|
coatomer protein complex, subunit gamma 2 |
| chr15_-_3976035 | 0.30 |
ENSDART00000168061
|
si:ch73-309g22.1
|
si:ch73-309g22.1 |
| chr3_-_53533128 | 0.29 |
ENSDART00000183591
|
notch3
|
notch 3 |
| chr15_+_31515976 | 0.29 |
ENSDART00000156471
|
tex26
|
testis expressed 26 |
| chr1_+_32521469 | 0.29 |
ENSDART00000113818
ENSDART00000152580 |
nlgn4a
|
neuroligin 4a |
| chr9_-_6372535 | 0.29 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr17_-_28705264 | 0.29 |
ENSDART00000076409
ENSDART00000189874 |
ap4s1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr19_+_32553874 | 0.29 |
ENSDART00000078197
|
heyl
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr13_-_33007781 | 0.29 |
ENSDART00000183671
ENSDART00000179859 |
rbm25a
|
RNA binding motif protein 25a |
| chr24_+_7782313 | 0.28 |
ENSDART00000111090
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr5_+_22791686 | 0.28 |
ENSDART00000014806
|
npas2
|
neuronal PAS domain protein 2 |
| chr22_-_4644484 | 0.28 |
ENSDART00000167748
|
fbn2b
|
fibrillin 2b |
| chr5_+_37785152 | 0.28 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr21_+_13353263 | 0.26 |
ENSDART00000114677
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr10_-_7785930 | 0.26 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr5_+_58492699 | 0.26 |
ENSDART00000181584
|
FQ378016.1
|
|
| chr21_+_9997418 | 0.26 |
ENSDART00000181454
ENSDART00000171579 |
herc7
|
hect domain and RLD 7 |
| chr14_+_14225048 | 0.24 |
ENSDART00000168749
|
nlgn3a
|
neuroligin 3a |
| chr19_+_30662529 | 0.24 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr14_+_5385855 | 0.24 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr20_-_23026223 | 0.23 |
ENSDART00000015755
|
rasl11b
|
RAS-like, family 11, member B |
| chr13_+_30874153 | 0.23 |
ENSDART00000112380
ENSDART00000189016 ENSDART00000134809 |
ercc6
|
excision repair cross-complementation group 6 |
| chr14_+_8127893 | 0.22 |
ENSDART00000169091
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr7_+_20019125 | 0.22 |
ENSDART00000186391
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr5_+_22970617 | 0.22 |
ENSDART00000192859
|
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr20_+_15982482 | 0.22 |
ENSDART00000020999
|
angptl1a
|
angiopoietin-like 1a |
| chr1_+_23784905 | 0.22 |
ENSDART00000171951
ENSDART00000188521 ENSDART00000183029 ENSDART00000187183 |
slit2
|
slit homolog 2 (Drosophila) |
| chr22_-_13165186 | 0.21 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr22_-_30935510 | 0.21 |
ENSDART00000133335
|
si:dkey-49n23.1
|
si:dkey-49n23.1 |
| chr11_+_30091155 | 0.20 |
ENSDART00000158691
|
si:ch211-161f7.2
|
si:ch211-161f7.2 |
| chr1_-_5746030 | 0.20 |
ENSDART00000150863
|
nrp2a
|
neuropilin 2a |
| chr4_+_5180650 | 0.20 |
ENSDART00000067390
|
fgf6b
|
fibroblast growth factor 6b |
| chr18_-_38087875 | 0.20 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr6_+_37308716 | 0.19 |
ENSDART00000085498
|
ptger3
|
prostaglandin E receptor 3 (subtype EP3) |
| chr5_-_67757188 | 0.19 |
ENSDART00000167168
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr4_-_14191434 | 0.19 |
ENSDART00000142374
ENSDART00000136730 |
pus7l
|
pseudouridylate synthase 7-like |
| chr17_-_37052622 | 0.19 |
ENSDART00000186408
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr4_-_14191717 | 0.18 |
ENSDART00000147928
|
pus7l
|
pseudouridylate synthase 7-like |
| chr9_-_23944470 | 0.18 |
ENSDART00000138754
|
col6a3
|
collagen, type VI, alpha 3 |
| chr16_-_24135508 | 0.18 |
ENSDART00000171819
ENSDART00000103176 |
bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr24_+_10310577 | 0.18 |
ENSDART00000141718
|
otulina
|
OTU deubiquitinase with linear linkage specificity a |
| chr17_-_37214196 | 0.17 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr9_+_6587364 | 0.17 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr9_+_6587056 | 0.16 |
ENSDART00000193421
|
fhl2a
|
four and a half LIM domains 2a |
| chr8_-_40251126 | 0.16 |
ENSDART00000180435
|
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr18_+_17786710 | 0.16 |
ENSDART00000190203
ENSDART00000187095 ENSDART00000083296 |
ZNF423
|
si:ch211-216l23.1 |
| chr23_-_18707418 | 0.16 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr10_-_43718914 | 0.16 |
ENSDART00000189277
|
cetn3
|
centrin 3 |
| chr2_+_33541928 | 0.15 |
ENSDART00000162852
|
BX548164.1
|
|
| chr18_-_23875219 | 0.15 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr18_-_38088099 | 0.15 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr23_-_30041065 | 0.15 |
ENSDART00000131209
ENSDART00000127192 |
ccdc187
|
coiled-coil domain containing 187 |
| chr9_-_27648683 | 0.15 |
ENSDART00000017292
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr13_-_13754091 | 0.15 |
ENSDART00000131255
|
ky
|
kyphoscoliosis peptidase |
| chr4_+_15954293 | 0.14 |
ENSDART00000132695
|
si:dkey-117n7.4
|
si:dkey-117n7.4 |
| chr2_+_30916188 | 0.14 |
ENSDART00000137012
|
myom1a
|
myomesin 1a (skelemin) |
| chr25_-_37465064 | 0.14 |
ENSDART00000186128
|
zgc:158366
|
zgc:158366 |
| chr25_+_13321307 | 0.14 |
ENSDART00000161284
|
si:ch211-194m7.5
|
si:ch211-194m7.5 |
| chr12_+_27536095 | 0.14 |
ENSDART00000013033
|
etv4
|
ets variant 4 |
| chr24_-_23671709 | 0.13 |
ENSDART00000164750
|
hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr9_-_30190513 | 0.13 |
ENSDART00000137083
|
impg2a
|
interphotoreceptor matrix proteoglycan 2a |
| chr4_-_28334464 | 0.13 |
ENSDART00000123617
|
celsr1a
|
cadherin EGF LAG seven-pass G-type receptor 1a |
| chr15_+_7054754 | 0.13 |
ENSDART00000149800
|
foxl2a
|
forkhead box L2a |
| chr8_+_25767610 | 0.13 |
ENSDART00000062406
|
cacna1sb
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, b |
| chr9_-_35069645 | 0.13 |
ENSDART00000122679
ENSDART00000077908 ENSDART00000077894 ENSDART00000125536 |
appb
|
amyloid beta (A4) precursor protein b |
| chr7_+_17356482 | 0.13 |
ENSDART00000063629
|
nitr3r.1l
|
novel immune-type receptor 3, related 1-like |
| chr17_-_37054959 | 0.13 |
ENSDART00000151921
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr16_+_10422836 | 0.12 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr7_-_33881275 | 0.12 |
ENSDART00000100102
|
rxfp3.3a1
|
relaxin/insulin-like family peptide receptor 3.3a1 |
| chr24_+_25919809 | 0.12 |
ENSDART00000006615
|
map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr15_+_3790556 | 0.12 |
ENSDART00000181467
|
RNF14 (1 of many)
|
ring finger protein 14 |
| chr9_+_31282161 | 0.12 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr7_+_25323742 | 0.12 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr5_-_28679135 | 0.11 |
ENSDART00000193585
|
tnc
|
tenascin C |
| chr15_-_4152582 | 0.11 |
ENSDART00000171942
|
si:dkey-83h2.5
|
si:dkey-83h2.5 |
| chr20_+_27240388 | 0.11 |
ENSDART00000123950
|
si:dkey-85n7.6
|
si:dkey-85n7.6 |
| chr4_-_28335184 | 0.11 |
ENSDART00000178149
ENSDART00000043737 |
celsr1a
|
cadherin EGF LAG seven-pass G-type receptor 1a |
| chr21_+_13908858 | 0.11 |
ENSDART00000148199
|
stxbp1a
|
syntaxin binding protein 1a |
| chr5_+_39087364 | 0.11 |
ENSDART00000004286
|
anxa3a
|
annexin A3a |
| chr25_+_20216159 | 0.11 |
ENSDART00000048642
|
tnnt2d
|
troponin T2d, cardiac |
| chr6_-_42971184 | 0.11 |
ENSDART00000014552
|
arl8ba
|
ADP-ribosylation factor-like 8Ba |
| chr12_+_27536270 | 0.10 |
ENSDART00000133719
|
etv4
|
ets variant 4 |
| chr16_-_44649053 | 0.10 |
ENSDART00000184807
|
CR925804.2
|
|
| chr4_-_16333944 | 0.10 |
ENSDART00000079523
|
epyc
|
epiphycan |
| chr23_-_26521970 | 0.10 |
ENSDART00000143712
|
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr1_+_52130213 | 0.10 |
ENSDART00000018817
|
rnf11a
|
ring finger protein 11a |
| chr6_-_52400896 | 0.10 |
ENSDART00000187624
|
mmp24
|
matrix metallopeptidase 24 |
| chr2_-_34555945 | 0.10 |
ENSDART00000056671
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr24_+_25069609 | 0.10 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr11_-_1948784 | 0.09 |
ENSDART00000082475
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr4_-_8902406 | 0.09 |
ENSDART00000192962
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr3_+_17456428 | 0.09 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr5_+_22264051 | 0.09 |
ENSDART00000143314
|
arhgap20b
|
Rho GTPase activating protein 20b |
| chr11_+_20056732 | 0.09 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr14_+_14224730 | 0.08 |
ENSDART00000180112
ENSDART00000184891 ENSDART00000174760 |
nlgn3a
|
neuroligin 3a |
| chr10_+_21867307 | 0.08 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr17_+_38262408 | 0.08 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of atf5a+atf5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.4 | 1.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 0.3 | 1.2 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.3 | 0.8 | GO:0060845 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.2 | 1.0 | GO:0031446 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 0.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 0.7 | GO:0034138 | toll-like receptor 2 signaling pathway(GO:0034134) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.2 | 1.0 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 0.5 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.1 | 0.3 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.3 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.1 | 0.3 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.3 | GO:0003249 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.4 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 0.7 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 0.8 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.7 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.2 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.7 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.3 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 1.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 1.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.0 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 0.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 2.0 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.7 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.2 | GO:0044058 | regulation of digestive system process(GO:0044058) |
| 0.0 | 0.2 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 1.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.1 | GO:0039022 | pronephric duct development(GO:0039022) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.3 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:0035992 | tendon formation(GO:0035992) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.0 | 0.7 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.3 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.3 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.5 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.4 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.6 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.3 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.0 | 0.4 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 2.8 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 0.1 | GO:0060959 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.0 | 0.4 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.3 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.6 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0001541 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.1 | GO:1903817 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) negative regulation of cation channel activity(GO:2001258) |
| 0.0 | 0.2 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.0 | 0.2 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.4 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.2 | GO:0038084 | vascular endothelial growth factor signaling pathway(GO:0038084) |
| 0.0 | 0.3 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.3 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.2 | 1.0 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 1.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 1.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.3 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 2.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 1.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 1.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 2.0 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 1.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 1.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.2 | 0.7 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 0.7 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.4 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 2.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.2 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.4 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 3.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 4.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |