Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
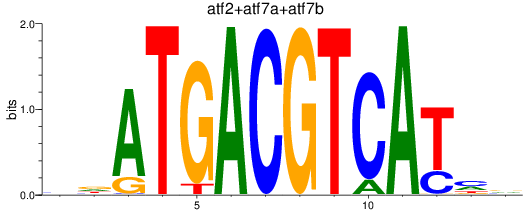
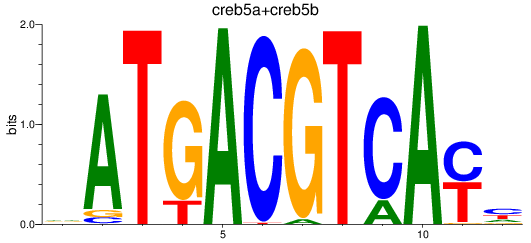
Results for atf2+atf7a+atf7b_creb5a+creb5b
Z-value: 0.88
Transcription factors associated with atf2+atf7a+atf7b_creb5a+creb5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf7a
|
ENSDARG00000011298 | activating transcription factor 7a |
|
atf2
|
ENSDARG00000023903 | activating transcription factor 2 |
|
atf7b
|
ENSDARG00000055481 | activating transcription factor 7b |
|
atf7b
|
ENSDARG00000114492 | activating transcription factor 7b |
|
atf7b
|
ENSDARG00000115171 | activating transcription factor 7b |
|
creb5b
|
ENSDARG00000070536 | cAMP responsive element binding protein 5b |
|
creb5a
|
ENSDARG00000099002 | cAMP responsive element binding protein 5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf7b | dr11_v1_chr6_-_39631164_39631181 | 0.97 | 7.4e-12 | Click! |
| creb5b | dr11_v1_chr16_-_20707742_20707742 | -0.59 | 9.6e-03 | Click! |
| creb5a | dr11_v1_chr19_-_19505167_19505167 | -0.56 | 1.5e-02 | Click! |
| atf7a | dr11_v1_chr23_-_27479558_27479558 | 0.50 | 3.6e-02 | Click! |
| atf2 | dr11_v1_chr9_+_2343096_2343172 | -0.29 | 2.5e-01 | Click! |
Activity profile of atf2+atf7a+atf7b_creb5a+creb5b motif
Sorted Z-values of atf2+atf7a+atf7b_creb5a+creb5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_35603637 | 8.12 |
ENSDART00000147278
|
plk3
|
polo-like kinase 3 (Drosophila) |
| chr16_-_29387215 | 3.85 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr11_-_44801968 | 3.47 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr2_+_12255568 | 3.33 |
ENSDART00000184164
ENSDART00000013454 |
prtfdc1
|
phosphoribosyl transferase domain containing 1 |
| chr7_-_51773166 | 3.25 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr25_-_6447835 | 2.89 |
ENSDART00000012820
|
snupn
|
snurportin 1 |
| chr14_+_30285613 | 2.89 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr24_-_19718077 | 2.69 |
ENSDART00000109107
ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr25_-_6448050 | 2.60 |
ENSDART00000180616
|
snupn
|
snurportin 1 |
| chr8_-_410199 | 2.39 |
ENSDART00000091177
ENSDART00000122979 ENSDART00000151331 ENSDART00000151155 |
trim36
|
tripartite motif containing 36 |
| chr18_+_8917766 | 2.36 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr8_-_410728 | 2.33 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr19_-_25772980 | 2.18 |
ENSDART00000052393
|
pard6gb
|
par-6 family cell polarity regulator gamma b |
| chr17_-_24866727 | 2.16 |
ENSDART00000027957
|
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr17_+_25187226 | 2.14 |
ENSDART00000148431
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr6_+_112579 | 2.12 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr17_-_24866964 | 2.07 |
ENSDART00000190601
ENSDART00000192547 |
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr15_+_20239141 | 1.90 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr12_-_17863467 | 1.90 |
ENSDART00000042006
|
baiap2l1a
|
BAI1-associated protein 2-like 1a |
| chr8_+_11687586 | 1.84 |
ENSDART00000146241
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr12_-_35505610 | 1.80 |
ENSDART00000105518
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr8_-_25846188 | 1.77 |
ENSDART00000128829
|
efhd2
|
EF-hand domain family, member D2 |
| chr2_-_10386738 | 1.73 |
ENSDART00000016369
|
wls
|
wntless Wnt ligand secretion mediator |
| chr7_-_69429561 | 1.73 |
ENSDART00000127351
|
atxn1l
|
ataxin 1-like |
| chr8_-_13678415 | 1.67 |
ENSDART00000134153
ENSDART00000143331 |
si:dkey-258f14.3
|
si:dkey-258f14.3 |
| chr18_-_8380090 | 1.66 |
ENSDART00000141581
ENSDART00000081143 |
sephs1
|
selenophosphate synthetase 1 |
| chr4_-_16628801 | 1.66 |
ENSDART00000040708
ENSDART00000064009 |
caprin2
|
caprin family member 2 |
| chr12_+_46869271 | 1.63 |
ENSDART00000166560
|
hpdl
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr25_-_34845302 | 1.60 |
ENSDART00000039485
|
gabarapl2
|
GABA(A) receptor-associated protein like 2 |
| chr1_+_10305611 | 1.59 |
ENSDART00000043881
|
zgc:77880
|
zgc:77880 |
| chr23_+_10805188 | 1.59 |
ENSDART00000035693
|
ppp4r2a
|
protein phosphatase 4, regulatory subunit 2a |
| chr17_+_15674052 | 1.59 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr4_+_76304911 | 1.59 |
ENSDART00000172734
ENSDART00000161850 |
si:ch73-389k6.1
|
si:ch73-389k6.1 |
| chr10_+_26990095 | 1.58 |
ENSDART00000064111
|
faub
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed b |
| chr12_-_41759686 | 1.56 |
ENSDART00000172175
ENSDART00000165152 |
ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr10_-_6588793 | 1.55 |
ENSDART00000163788
|
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr7_-_26532089 | 1.54 |
ENSDART00000121698
|
senp3b
|
SUMO1/sentrin/SMT3 specific peptidase 3b |
| chr9_+_34148714 | 1.54 |
ENSDART00000078051
|
gpr161
|
G protein-coupled receptor 161 |
| chr11_+_42474694 | 1.52 |
ENSDART00000056048
ENSDART00000184710 |
si:ch1073-165f9.2
|
si:ch1073-165f9.2 |
| chr15_-_20412286 | 1.47 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr14_+_1170968 | 1.47 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr9_-_6502491 | 1.45 |
ENSDART00000102672
|
nck2a
|
NCK adaptor protein 2a |
| chr6_-_29049022 | 1.43 |
ENSDART00000190309
|
evi5b
|
ecotropic viral integration site 5b |
| chr3_-_49514874 | 1.41 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr19_+_42227400 | 1.37 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr3_+_53116172 | 1.32 |
ENSDART00000115117
|
brd4
|
bromodomain containing 4 |
| chr23_-_29357764 | 1.31 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr7_+_70338270 | 1.30 |
ENSDART00000065234
|
gba3
|
glucosidase, beta, acid 3 (gene/pseudogene) |
| chr9_-_28939181 | 1.29 |
ENSDART00000101276
ENSDART00000135334 |
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr12_+_33320884 | 1.25 |
ENSDART00000188988
|
csnk1db
|
casein kinase 1, delta b |
| chr14_+_30413312 | 1.24 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr5_+_30089715 | 1.24 |
ENSDART00000078114
|
timm8b
|
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr15_+_34963316 | 1.23 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr8_+_48965767 | 1.22 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr23_-_27505825 | 1.22 |
ENSDART00000137229
ENSDART00000013797 |
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr6_+_21227621 | 1.22 |
ENSDART00000193583
|
prkca
|
protein kinase C, alpha |
| chr2_+_9560740 | 1.22 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr20_-_3319642 | 1.21 |
ENSDART00000186743
ENSDART00000123096 |
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr4_-_12930086 | 1.21 |
ENSDART00000013604
|
lemd3
|
LEM domain containing 3 |
| chr15_-_1765098 | 1.18 |
ENSDART00000149980
ENSDART00000093074 |
bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr23_+_34990693 | 1.17 |
ENSDART00000013449
|
CHST13
|
si:ch211-236h17.3 |
| chr16_-_9869056 | 1.16 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr23_+_29358188 | 1.15 |
ENSDART00000189242
|
tardbpl
|
TAR DNA binding protein, like |
| chr22_-_38934989 | 1.14 |
ENSDART00000008365
|
ncbp2
|
nuclear cap binding protein subunit 2 |
| chr23_-_25686894 | 1.14 |
ENSDART00000181420
ENSDART00000088208 |
lrp1ab
|
low density lipoprotein receptor-related protein 1Ab |
| chr2_-_17114852 | 1.13 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr5_+_26921033 | 1.10 |
ENSDART00000051483
|
tm2d2
|
TM2 domain containing 2 |
| chr25_+_3104959 | 1.10 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr18_+_8912113 | 1.10 |
ENSDART00000147467
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr7_-_33868903 | 1.10 |
ENSDART00000173500
ENSDART00000178746 |
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr13_-_49444636 | 1.09 |
ENSDART00000136991
|
irf2bp2a
|
interferon regulatory factor 2 binding protein 2a |
| chr5_+_22133153 | 1.08 |
ENSDART00000016214
|
msna
|
moesin a |
| chr9_+_25840720 | 1.08 |
ENSDART00000024572
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr2_-_17115256 | 1.08 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr5_-_30145939 | 1.06 |
ENSDART00000086795
|
zbtb44
|
zinc finger and BTB domain containing 44 |
| chr25_-_37084032 | 1.06 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr5_-_37959874 | 1.05 |
ENSDART00000031719
|
mpzl2b
|
myelin protein zero-like 2b |
| chr19_+_42660158 | 1.04 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr4_-_73488406 | 1.04 |
ENSDART00000115002
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr23_-_43424510 | 1.03 |
ENSDART00000055564
|
rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr4_+_73051901 | 1.03 |
ENSDART00000174219
|
zgc:152938
|
zgc:152938 |
| chr22_+_1940595 | 1.03 |
ENSDART00000163506
|
znf1167
|
zinc finger protein 1167 |
| chr13_+_51710725 | 1.02 |
ENSDART00000163741
|
pwwp2b
|
PWWP domain containing 2B |
| chr16_+_14201401 | 1.01 |
ENSDART00000113679
|
dap3
|
death associated protein 3 |
| chr23_-_27506161 | 1.00 |
ENSDART00000145007
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr2_-_4032732 | 1.00 |
ENSDART00000158335
|
rab18b
|
RAB18B, member RAS oncogene family |
| chr15_+_807897 | 1.00 |
ENSDART00000153549
|
si:dkey-7i4.8
|
si:dkey-7i4.8 |
| chr23_-_35396845 | 0.99 |
ENSDART00000142038
ENSDART00000049373 ENSDART00000181978 ENSDART00000171357 |
cmtr1
|
cap methyltransferase 1 |
| chr19_+_26923110 | 0.97 |
ENSDART00000149988
|
nelfe
|
negative elongation factor complex member E |
| chr18_-_20444296 | 0.96 |
ENSDART00000132993
|
kif23
|
kinesin family member 23 |
| chr5_-_19444930 | 0.95 |
ENSDART00000136259
ENSDART00000188499 |
kctd10
|
potassium channel tetramerization domain containing 10 |
| chr23_-_1571682 | 0.94 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr17_+_23554932 | 0.94 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr15_-_43995028 | 0.93 |
ENSDART00000172485
ENSDART00000186320 |
nlrc3l
|
NLR family, CARD domain containing 3-like |
| chr7_-_19999152 | 0.93 |
ENSDART00000173881
ENSDART00000100798 |
trip6
|
thyroid hormone receptor interactor 6 |
| chr20_+_39223235 | 0.93 |
ENSDART00000132132
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr18_+_8912536 | 0.92 |
ENSDART00000134827
ENSDART00000061904 |
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr23_-_17509656 | 0.91 |
ENSDART00000148423
|
dnajc5ab
|
DnaJ (Hsp40) homolog, subfamily C, member 5ab |
| chr16_-_41667101 | 0.91 |
ENSDART00000084528
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr24_+_26329018 | 0.90 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr8_-_38317914 | 0.90 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr21_-_25395223 | 0.90 |
ENSDART00000016219
|
ppme1
|
protein phosphatase methylesterase 1 |
| chr17_-_8674208 | 0.90 |
ENSDART00000149201
|
ctbp2a
|
C-terminal binding protein 2a |
| chr24_-_18876877 | 0.90 |
ENSDART00000186269
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr7_+_28612671 | 0.88 |
ENSDART00000019991
|
slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr4_+_77141970 | 0.88 |
ENSDART00000174209
|
si:dkey-172k15.11
|
si:dkey-172k15.11 |
| chr4_+_76466751 | 0.87 |
ENSDART00000164709
|
zgc:153116
|
zgc:153116 |
| chr16_+_25137483 | 0.84 |
ENSDART00000155666
|
znf576.1
|
zinc finger protein 576, tandem duplicate 1 |
| chr23_+_4709607 | 0.83 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr1_-_23595779 | 0.83 |
ENSDART00000134860
ENSDART00000138852 |
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr11_+_25296366 | 0.82 |
ENSDART00000065949
|
top1l
|
DNA topoisomerase I, like |
| chr7_+_34549198 | 0.82 |
ENSDART00000173784
|
fhod1
|
formin homology 2 domain containing 1 |
| chr2_-_43739559 | 0.81 |
ENSDART00000138947
|
kif5ba
|
kinesin family member 5B, a |
| chr7_-_19998723 | 0.81 |
ENSDART00000173458
|
trip6
|
thyroid hormone receptor interactor 6 |
| chr7_+_7552008 | 0.81 |
ENSDART00000173018
ENSDART00000049311 |
clcn3
|
chloride channel 3 |
| chr2_-_37477654 | 0.80 |
ENSDART00000193921
|
dapk3
|
death-associated protein kinase 3 |
| chr17_-_8673567 | 0.80 |
ENSDART00000192714
ENSDART00000012546 |
ctbp2a
|
C-terminal binding protein 2a |
| chr13_-_17464362 | 0.80 |
ENSDART00000145499
|
lrmda
|
leucine rich melanocyte differentiation associated |
| chr24_+_26328787 | 0.80 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr24_-_37326236 | 0.80 |
ENSDART00000003789
|
pdpk1b
|
3-phosphoinositide dependent protein kinase 1b |
| chr2_-_56635744 | 0.79 |
ENSDART00000167790
ENSDART00000168160 |
pip5k1cb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b |
| chr20_-_37933237 | 0.79 |
ENSDART00000142567
ENSDART00000036371 ENSDART00000061445 |
angel2
|
angel homolog 2 (Drosophila) |
| chr21_-_45382112 | 0.77 |
ENSDART00000151029
ENSDART00000151335 ENSDART00000151687 ENSDART00000075438 |
cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr19_+_26923274 | 0.77 |
ENSDART00000148439
ENSDART00000148877 |
nelfe
|
negative elongation factor complex member E |
| chr8_-_1267247 | 0.76 |
ENSDART00000150064
|
cdc14b
|
cell division cycle 14B |
| chr7_+_34549377 | 0.76 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr21_+_19547806 | 0.76 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr4_+_73651053 | 0.75 |
ENSDART00000150532
ENSDART00000172536 |
znf989
|
zinc finger protein 989 |
| chr25_-_25646806 | 0.75 |
ENSDART00000089066
|
tbc1d2b
|
TBC1 domain family, member 2B |
| chr23_+_1661743 | 0.75 |
ENSDART00000044776
|
stxbp3
|
syntaxin binding protein 3 |
| chr21_-_23307653 | 0.75 |
ENSDART00000140284
ENSDART00000134103 |
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr14_+_30413758 | 0.74 |
ENSDART00000092953
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr10_+_28306749 | 0.74 |
ENSDART00000142016
|
ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr12_+_33320504 | 0.74 |
ENSDART00000021491
|
csnk1db
|
casein kinase 1, delta b |
| chr6_+_27667359 | 0.73 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr6_+_13232934 | 0.72 |
ENSDART00000089725
|
ino80db
|
INO80 complex subunit Db |
| chr4_-_1839192 | 0.72 |
ENSDART00000003790
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr23_-_29667544 | 0.71 |
ENSDART00000059339
|
clstn1
|
calsyntenin 1 |
| chr5_+_1965296 | 0.71 |
ENSDART00000156224
|
dhx33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr24_-_18877118 | 0.70 |
ENSDART00000092783
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr24_-_16979728 | 0.70 |
ENSDART00000005331
|
klhl15
|
kelch-like family member 15 |
| chr7_+_15659280 | 0.70 |
ENSDART00000173414
|
mef2ab
|
myocyte enhancer factor 2ab |
| chr2_-_43739740 | 0.69 |
ENSDART00000113849
|
kif5ba
|
kinesin family member 5B, a |
| chr8_-_31417139 | 0.69 |
ENSDART00000180204
|
znf131
|
zinc finger protein 131 |
| chr22_-_718615 | 0.69 |
ENSDART00000149320
|
arl8a
|
ADP-ribosylation factor-like 8A |
| chr20_-_34801181 | 0.68 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr2_-_55779927 | 0.68 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr13_-_25774183 | 0.68 |
ENSDART00000046981
|
pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr16_+_27614989 | 0.68 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr14_-_38946808 | 0.66 |
ENSDART00000139293
|
gla
|
galactosidase, alpha |
| chr17_-_31212420 | 0.66 |
ENSDART00000086511
|
rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr6_+_6802582 | 0.65 |
ENSDART00000189422
|
dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr4_+_9478500 | 0.65 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr20_+_23947004 | 0.65 |
ENSDART00000144195
|
casp8ap2
|
caspase 8 associated protein 2 |
| chr4_+_76305357 | 0.65 |
ENSDART00000160326
|
si:ch73-389k6.1
|
si:ch73-389k6.1 |
| chr3_+_25907266 | 0.65 |
ENSDART00000170324
ENSDART00000192633 |
tom1
|
target of myb1 membrane trafficking protein |
| chr18_+_14633974 | 0.65 |
ENSDART00000133834
|
vps9d1
|
VPS9 domain containing 1 |
| chr18_+_27598755 | 0.65 |
ENSDART00000193808
|
cd82b
|
CD82 molecule b |
| chr1_+_19538299 | 0.64 |
ENSDART00000109416
|
smc2
|
structural maintenance of chromosomes 2 |
| chr20_-_44576949 | 0.64 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr1_+_49878000 | 0.64 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr23_+_29357790 | 0.63 |
ENSDART00000139223
ENSDART00000141928 |
tardbpl
|
TAR DNA binding protein, like |
| chr24_-_37338162 | 0.63 |
ENSDART00000056303
|
tsr3
|
TSR3, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr22_+_38935060 | 0.62 |
ENSDART00000183732
ENSDART00000130055 |
sirt7
|
sirtuin 7 |
| chr23_+_23183449 | 0.62 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr9_-_6501924 | 0.62 |
ENSDART00000168143
|
nck2a
|
NCK adaptor protein 2a |
| chr24_+_37338169 | 0.62 |
ENSDART00000141771
|
gnptg
|
N-acetylglucosamine-1-phosphate transferase, gamma subunit |
| chr11_-_575786 | 0.61 |
ENSDART00000019997
|
mkrn2
|
makorin, ring finger protein, 2 |
| chr12_+_36109507 | 0.60 |
ENSDART00000175409
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr20_+_36812368 | 0.60 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr1_-_29061285 | 0.60 |
ENSDART00000053933
ENSDART00000142350 ENSDART00000192615 |
gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr23_-_10722664 | 0.59 |
ENSDART00000146526
ENSDART00000129022 ENSDART00000104985 |
foxp1a
|
forkhead box P1a |
| chr11_+_24314148 | 0.58 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr10_+_44903676 | 0.58 |
ENSDART00000158553
|
zgc:114173
|
zgc:114173 |
| chr21_-_36571804 | 0.58 |
ENSDART00000138129
|
wwc1
|
WW and C2 domain containing 1 |
| chr22_+_1517318 | 0.56 |
ENSDART00000160406
|
si:ch211-255f4.5
|
si:ch211-255f4.5 |
| chr5_+_28259655 | 0.56 |
ENSDART00000087684
|
ncaph
|
non-SMC condensin I complex, subunit H |
| chr12_+_38807604 | 0.56 |
ENSDART00000155563
|
abca5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr22_+_1947494 | 0.56 |
ENSDART00000159121
|
si:dkey-15h8.15
|
si:dkey-15h8.15 |
| chr1_+_52792439 | 0.56 |
ENSDART00000123972
|
smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr13_-_40754499 | 0.56 |
ENSDART00000111641
ENSDART00000159255 |
morn4
|
MORN repeat containing 4 |
| chr5_+_56277866 | 0.56 |
ENSDART00000170610
ENSDART00000028854 ENSDART00000148749 |
aatf
|
apoptosis antagonizing transcription factor |
| chr14_+_16083818 | 0.55 |
ENSDART00000168462
|
rnf103
|
ring finger protein 103 |
| chr8_+_1839695 | 0.55 |
ENSDART00000148254
ENSDART00000143473 |
snap29
|
synaptosomal-associated protein 29 |
| chr4_-_1839352 | 0.55 |
ENSDART00000189215
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr8_+_29742237 | 0.54 |
ENSDART00000133955
ENSDART00000020621 |
mapk4
|
mitogen-activated protein kinase 4 |
| chr21_-_44772738 | 0.54 |
ENSDART00000026178
|
kif4
|
kinesin family member 4 |
| chr18_-_12451772 | 0.53 |
ENSDART00000175083
|
si:ch211-1e14.1
|
si:ch211-1e14.1 |
| chr6_-_25952848 | 0.53 |
ENSDART00000076997
ENSDART00000148748 |
lmo4b
|
LIM domain only 4b |
| chr1_-_18592068 | 0.53 |
ENSDART00000082063
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr18_+_910992 | 0.53 |
ENSDART00000161206
ENSDART00000167229 |
pkma
|
pyruvate kinase M1/2a |
| chr17_-_5610514 | 0.53 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr6_-_14010554 | 0.52 |
ENSDART00000004656
|
zgc:92027
|
zgc:92027 |
| chr14_+_20351 | 0.52 |
ENSDART00000051893
|
stx18
|
syntaxin 18 |
| chr15_+_23443665 | 0.52 |
ENSDART00000059369
|
phykpl
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr20_+_46699021 | 0.51 |
ENSDART00000167398
ENSDART00000029894 |
wdr26b
|
WD repeat domain 26b |
| chr15_-_704408 | 0.51 |
ENSDART00000156200
ENSDART00000166404 ENSDART00000131040 |
zgc:174574
|
zgc:174574 |
| chr10_-_1276046 | 0.51 |
ENSDART00000169779
|
pdlim5b
|
PDZ and LIM domain 5b |
| chr8_-_13735572 | 0.51 |
ENSDART00000139642
|
si:dkey-258f14.7
|
si:dkey-258f14.7 |
| chr25_-_12906872 | 0.50 |
ENSDART00000165156
ENSDART00000167449 |
sept15
|
septin 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of atf2+atf7a+atf7b_creb5a+creb5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.1 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 1.4 | 5.5 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 1.1 | 4.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.8 | 3.3 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.6 | 1.7 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.6 | 1.1 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.6 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.6 | 1.7 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.4 | 1.3 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.4 | 1.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.4 | 1.1 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.4 | 1.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 1.3 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.3 | 1.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 | 1.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.3 | 1.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.3 | 5.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 2.0 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 1.7 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.2 | 0.9 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 1.9 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 1.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.2 | 1.7 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.2 | 1.3 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.2 | 1.7 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.2 | 0.9 | GO:0014005 | microglia development(GO:0014005) |
| 0.2 | 2.2 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 0.9 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.2 | 1.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 2.2 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.2 | 0.5 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.2 | 1.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 1.2 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 1.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.6 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.1 | 0.9 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.1 | 0.6 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.5 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 1.8 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.7 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.8 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.5 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.4 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 1.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 1.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.4 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.6 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 0.3 | GO:0051103 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 1.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.3 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 0.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.8 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 1.4 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.5 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.5 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.2 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 2.8 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.1 | 1.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.9 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.3 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.3 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.3 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.6 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 5.7 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.1 | 0.3 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.1 | 2.7 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 0.3 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.8 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 1.6 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.5 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.3 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 2.6 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.2 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.5 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.2 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 3.1 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.6 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 1.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.8 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.7 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 2.2 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 1.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.9 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.3 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 1.1 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 1.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 1.5 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.0 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.0 | 0.2 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.0 | 2.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.1 | GO:0034334 | cell junction maintenance(GO:0034331) adherens junction maintenance(GO:0034334) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.3 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.2 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.0 | 0.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.5 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.3 | 1.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.2 | 1.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 1.1 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.2 | 2.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 5.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 3.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.8 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 8.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.4 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 2.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.4 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 1.8 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.3 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.6 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.3 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 4.6 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 2.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 1.4 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 1.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.1 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 4.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.2 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.3 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.0 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.2 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.5 | 1.6 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.4 | 2.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.4 | 2.6 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.4 | 1.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.3 | 5.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.3 | 1.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 2.1 | GO:0046625 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.2 | 0.7 | GO:0015925 | alpha-galactosidase activity(GO:0004557) galactosidase activity(GO:0015925) |
| 0.2 | 0.7 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.2 | 1.3 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.2 | 2.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 0.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 3.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 2.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.4 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 2.6 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.8 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 2.1 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 1.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.4 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 1.2 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 0.6 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 5.5 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 1.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.2 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.1 | 0.8 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.5 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.2 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.6 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 2.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.5 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.7 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 1.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.4 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 2.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.8 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 2.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.6 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 3.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.0 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 8.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 8.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 7.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 2.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 6.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 1.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 2.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 4.4 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.6 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 3.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |