Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
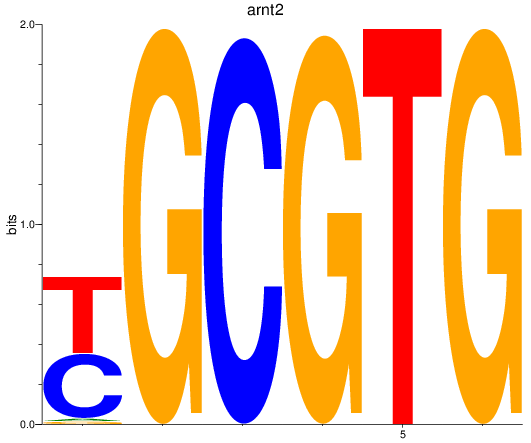
Results for arnt2
Z-value: 1.70
Transcription factors associated with arnt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arnt2
|
ENSDARG00000103697 | aryl-hydrocarbon receptor nuclear translocator 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arnt2 | dr11_v1_chr7_+_10701938_10701938 | 0.96 | 1.8e-10 | Click! |
Activity profile of arnt2 motif
Sorted Z-values of arnt2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_10152092 | 5.51 |
ENSDART00000066053
|
cbx2
|
chromobox homolog 2 (Drosophila Pc class) |
| chr1_+_49814461 | 5.03 |
ENSDART00000132405
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr4_+_9279784 | 4.32 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr10_-_25217347 | 4.01 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr3_-_1283247 | 3.94 |
ENSDART00000149814
|
tcf20
|
transcription factor 20 |
| chr20_-_48898371 | 3.86 |
ENSDART00000170617
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr12_+_38807604 | 3.77 |
ENSDART00000155563
|
abca5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr2_-_21438492 | 3.69 |
ENSDART00000046098
|
plcd1b
|
phospholipase C, delta 1b |
| chr8_-_18899427 | 3.51 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr5_+_25733774 | 3.46 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr10_+_29963518 | 3.40 |
ENSDART00000011317
ENSDART00000099964 ENSDART00000182990 ENSDART00000113912 |
ntm
|
neurotrimin |
| chr2_-_21820697 | 3.17 |
ENSDART00000135230
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr21_-_30082414 | 3.15 |
ENSDART00000157307
ENSDART00000155188 |
ccnjl
|
cyclin J-like |
| chr15_-_33734105 | 3.13 |
ENSDART00000172729
ENSDART00000172045 |
trmt9b
|
tRNA methyltransferase 9B |
| chr11_-_34577034 | 3.13 |
ENSDART00000133302
ENSDART00000184367 |
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr24_-_42072886 | 3.04 |
ENSDART00000171389
|
CABZ01095370.1
|
|
| chr20_-_182841 | 3.03 |
ENSDART00000064546
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr6_-_53144336 | 2.98 |
ENSDART00000154429
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr23_-_26227805 | 2.91 |
ENSDART00000158082
|
BX927204.1
|
|
| chr22_-_718615 | 2.89 |
ENSDART00000149320
|
arl8a
|
ADP-ribosylation factor-like 8A |
| chr4_-_77561679 | 2.88 |
ENSDART00000180809
|
AL935186.9
|
|
| chr18_-_20608025 | 2.88 |
ENSDART00000090156
ENSDART00000151980 |
bcl2l13
|
BCL2 like 13 |
| chr20_+_27712714 | 2.87 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr22_+_10781894 | 2.86 |
ENSDART00000081183
|
enc3
|
ectodermal-neural cortex 3 |
| chr4_+_2482046 | 2.84 |
ENSDART00000103371
|
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr7_+_10701938 | 2.81 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr21_-_13856689 | 2.81 |
ENSDART00000102197
|
fam129ba
|
family with sequence similarity 129, member Ba |
| chr16_-_34195002 | 2.79 |
ENSDART00000054026
|
rcc1
|
regulator of chromosome condensation 1 |
| chr8_+_50190742 | 2.73 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr23_-_26228077 | 2.71 |
ENSDART00000162423
|
BX927204.1
|
|
| chr13_-_15929402 | 2.71 |
ENSDART00000090273
|
ttl
|
tubulin tyrosine ligase |
| chr20_+_38543542 | 2.71 |
ENSDART00000145254
|
gtf3c2
|
general transcription factor IIIC, polypeptide 2, beta |
| chr6_-_53143667 | 2.66 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr21_-_4849029 | 2.64 |
ENSDART00000168930
ENSDART00000151019 |
notch1a
|
notch 1a |
| chr14_+_24840669 | 2.61 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr3_-_60589292 | 2.61 |
ENSDART00000157822
|
jmjd6
|
jumonji domain containing 6 |
| chr17_-_12249990 | 2.60 |
ENSDART00000177889
ENSDART00000155545 |
ahctf1
|
AT hook containing transcription factor 1 |
| chr17_+_19630272 | 2.59 |
ENSDART00000104895
|
rgs7a
|
regulator of G protein signaling 7a |
| chr8_+_2642384 | 2.58 |
ENSDART00000143242
|
naif1
|
nuclear apoptosis inducing factor 1 |
| chr7_-_60351876 | 2.58 |
ENSDART00000098563
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr17_+_34805897 | 2.53 |
ENSDART00000137090
ENSDART00000077626 |
id2a
|
inhibitor of DNA binding 2a |
| chr15_-_4616816 | 2.51 |
ENSDART00000160191
ENSDART00000161721 ENSDART00000144949 |
eif4h
|
eukaryotic translation initiation factor 4h |
| chr13_+_33268657 | 2.50 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr5_-_22130937 | 2.46 |
ENSDART00000138606
|
las1l
|
LAS1-like, ribosome biogenesis factor |
| chr15_-_16076399 | 2.46 |
ENSDART00000135658
ENSDART00000133755 ENSDART00000080413 |
srsf1a
|
serine/arginine-rich splicing factor 1a |
| chr18_-_46684352 | 2.45 |
ENSDART00000167520
|
pid1
|
phosphotyrosine interaction domain containing 1 |
| chr7_-_60351537 | 2.44 |
ENSDART00000159875
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr10_+_5268054 | 2.40 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr4_-_15003854 | 2.38 |
ENSDART00000134701
ENSDART00000002401 |
klhdc10
|
kelch domain containing 10 |
| chr13_-_24826607 | 2.37 |
ENSDART00000087786
ENSDART00000186951 |
slka
|
STE20-like kinase a |
| chr16_-_7793457 | 2.36 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr9_-_34945566 | 2.36 |
ENSDART00000131908
ENSDART00000059861 |
dcun1d2a
|
DCN1, defective in cullin neddylation 1, domain containing 2a |
| chr3_-_25054002 | 2.36 |
ENSDART00000086768
|
ep300b
|
E1A binding protein p300 b |
| chr19_-_30800004 | 2.31 |
ENSDART00000128560
ENSDART00000045504 ENSDART00000125893 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr13_-_45022301 | 2.30 |
ENSDART00000183589
ENSDART00000125633 ENSDART00000074787 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr22_-_21150845 | 2.30 |
ENSDART00000027345
|
tmem59l
|
transmembrane protein 59-like |
| chr1_+_49814942 | 2.29 |
ENSDART00000164936
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr22_+_25086942 | 2.27 |
ENSDART00000061117
|
rrbp1b
|
ribosome binding protein 1b |
| chr4_-_12930086 | 2.27 |
ENSDART00000013604
|
lemd3
|
LEM domain containing 3 |
| chr2_-_32594777 | 2.22 |
ENSDART00000134125
|
fastk
|
Fas-activated serine/threonine kinase |
| chr7_+_10701770 | 2.21 |
ENSDART00000167323
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr16_+_54641230 | 2.21 |
ENSDART00000157641
ENSDART00000159540 |
fbxo43
|
F-box protein 43 |
| chr15_-_34865952 | 2.18 |
ENSDART00000186868
|
sh3bp5la
|
SH3-binding domain protein 5-like, a |
| chr15_+_34963316 | 2.17 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr14_-_24110707 | 2.15 |
ENSDART00000133522
ENSDART00000123152 |
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr9_-_12269847 | 2.14 |
ENSDART00000136558
ENSDART00000144734 ENSDART00000131766 ENSDART00000032344 |
nup35
|
nucleoporin 35 |
| chr8_+_23861461 | 2.14 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr10_+_23060391 | 2.13 |
ENSDART00000079711
|
slc25a1a
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1a |
| chr19_+_41006975 | 2.12 |
ENSDART00000138555
ENSDART00000049842 |
casd1
|
CAS1 domain containing 1 |
| chr14_+_50918769 | 2.10 |
ENSDART00000146918
|
rnf44
|
ring finger protein 44 |
| chr22_+_25086567 | 2.09 |
ENSDART00000192114
ENSDART00000177284 ENSDART00000180296 ENSDART00000190384 |
rrbp1b
|
ribosome binding protein 1b |
| chr12_-_24832297 | 2.08 |
ENSDART00000066317
|
foxn2b
|
forkhead box N2b |
| chr3_+_19665319 | 2.06 |
ENSDART00000007857
ENSDART00000193509 |
mettl2a
|
methyltransferase like 2A |
| chr15_-_8856391 | 2.06 |
ENSDART00000008273
|
rab4b
|
RAB4B, member RAS oncogene family |
| chr14_+_8947282 | 2.04 |
ENSDART00000047993
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr20_+_27713210 | 2.04 |
ENSDART00000132222
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr10_+_8197827 | 2.03 |
ENSDART00000026244
|
mtrex
|
Mtr4 exosome RNA helicase |
| chr1_-_55248496 | 2.02 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr17_+_19630068 | 2.01 |
ENSDART00000182619
|
rgs7a
|
regulator of G protein signaling 7a |
| chr1_-_40519340 | 1.99 |
ENSDART00000114659
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr5_+_27583445 | 1.98 |
ENSDART00000136488
|
zmat4a
|
zinc finger, matrin-type 4a |
| chr19_-_43757568 | 1.98 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr12_-_48943467 | 1.97 |
ENSDART00000191829
|
CABZ01092907.1
|
|
| chr1_-_53685090 | 1.97 |
ENSDART00000162751
|
xpo1a
|
exportin 1 (CRM1 homolog, yeast) a |
| chr25_-_34845302 | 1.97 |
ENSDART00000039485
|
gabarapl2
|
GABA(A) receptor-associated protein like 2 |
| chr15_-_34866219 | 1.96 |
ENSDART00000099723
|
sh3bp5la
|
SH3-binding domain protein 5-like, a |
| chr18_-_8380090 | 1.96 |
ENSDART00000141581
ENSDART00000081143 |
sephs1
|
selenophosphate synthetase 1 |
| chr3_-_34136368 | 1.96 |
ENSDART00000136900
ENSDART00000186125 |
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr13_-_45022527 | 1.94 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr11_+_14321113 | 1.94 |
ENSDART00000039822
ENSDART00000137347 ENSDART00000132997 |
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr15_+_30126971 | 1.94 |
ENSDART00000100214
|
nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr13_-_15928934 | 1.89 |
ENSDART00000142732
|
ttl
|
tubulin tyrosine ligase |
| chr8_-_19266325 | 1.89 |
ENSDART00000036148
ENSDART00000137994 |
zgc:77486
|
zgc:77486 |
| chr6_+_38626684 | 1.88 |
ENSDART00000086533
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr2_+_38271392 | 1.87 |
ENSDART00000042100
|
homeza
|
homeobox and leucine zipper encoding a |
| chr10_+_11260170 | 1.85 |
ENSDART00000155742
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr15_+_29025090 | 1.83 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr19_+_46113828 | 1.82 |
ENSDART00000159331
ENSDART00000161826 |
rbm24a
|
RNA binding motif protein 24a |
| chr15_+_28175638 | 1.81 |
ENSDART00000037119
|
slc46a1
|
solute carrier family 46 (folate transporter), member 1 |
| chr25_-_25575717 | 1.81 |
ENSDART00000067138
|
hic1l
|
hypermethylated in cancer 1 like |
| chr21_+_6114709 | 1.80 |
ENSDART00000065858
|
fpgs
|
folylpolyglutamate synthase |
| chr7_-_55633475 | 1.80 |
ENSDART00000149478
|
galns
|
galactosamine (N-acetyl)-6-sulfatase |
| chr15_+_17406920 | 1.76 |
ENSDART00000081059
|
rps6kb1b
|
ribosomal protein S6 kinase b, polypeptide 1b |
| chr23_+_38159715 | 1.75 |
ENSDART00000137969
|
zgc:112994
|
zgc:112994 |
| chr2_-_2096055 | 1.74 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr7_+_17907883 | 1.74 |
ENSDART00000147564
|
mta2
|
metastasis associated 1 family, member 2 |
| chr19_+_42660158 | 1.74 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr8_-_5267442 | 1.72 |
ENSDART00000155816
|
ppp2r2aa
|
protein phosphatase 2, regulatory subunit B, alpha a |
| chr10_+_29138021 | 1.72 |
ENSDART00000025227
ENSDART00000123033 ENSDART00000034242 |
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr22_-_20695237 | 1.71 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr23_+_35426404 | 1.71 |
ENSDART00000164658
|
si:ch211-225h24.2
|
si:ch211-225h24.2 |
| chr6_+_38626926 | 1.71 |
ENSDART00000190339
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr14_+_31509922 | 1.70 |
ENSDART00000124499
|
hprt1
|
hypoxanthine phosphoribosyltransferase 1 |
| chr8_+_21280360 | 1.70 |
ENSDART00000144488
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr15_-_43978141 | 1.68 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr20_-_31743817 | 1.68 |
ENSDART00000137679
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr6_+_296130 | 1.68 |
ENSDART00000073985
|
rbfox2
|
RNA binding fox-1 homolog 2 |
| chr12_-_25150239 | 1.68 |
ENSDART00000038415
ENSDART00000135368 |
rhoq
|
ras homolog family member Q |
| chr6_+_33537267 | 1.67 |
ENSDART00000040334
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr25_-_36263115 | 1.66 |
ENSDART00000143046
ENSDART00000139002 |
dus2
|
dihydrouridine synthase 2 |
| chr14_+_14806851 | 1.65 |
ENSDART00000169235
|
fhdc2
|
FH2 domain containing 2 |
| chr17_+_6538733 | 1.65 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr17_+_46387086 | 1.65 |
ENSDART00000157079
|
si:dkey-206p8.1
|
si:dkey-206p8.1 |
| chr8_-_22542467 | 1.65 |
ENSDART00000182588
ENSDART00000134542 |
csde1
|
cold shock domain containing E1, RNA-binding |
| chr17_+_15535501 | 1.64 |
ENSDART00000002932
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr24_-_38083378 | 1.64 |
ENSDART00000056381
|
crp2
|
C-reactive protein 2 |
| chr23_+_36616717 | 1.63 |
ENSDART00000042701
ENSDART00000192980 |
pip4k2ca
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma a |
| chr13_+_40815012 | 1.63 |
ENSDART00000016960
|
prkg1a
|
protein kinase, cGMP-dependent, type Ia |
| chr4_+_25181572 | 1.62 |
ENSDART00000078529
ENSDART00000136643 |
kin
|
Kin17 DNA and RNA binding protein |
| chr9_+_38588081 | 1.61 |
ENSDART00000031127
ENSDART00000131784 |
snx4
|
sorting nexin 4 |
| chr3_+_34149337 | 1.61 |
ENSDART00000006091
|
carm1
|
coactivator-associated arginine methyltransferase 1 |
| chr6_-_37744430 | 1.60 |
ENSDART00000150177
ENSDART00000149722 |
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr3_-_34136778 | 1.60 |
ENSDART00000131951
|
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr1_+_36772348 | 1.60 |
ENSDART00000109314
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr16_+_35535171 | 1.59 |
ENSDART00000167001
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr22_-_11054244 | 1.59 |
ENSDART00000105823
|
insrb
|
insulin receptor b |
| chr2_-_15040345 | 1.59 |
ENSDART00000109657
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr18_-_38271298 | 1.58 |
ENSDART00000143016
|
caprin1b
|
cell cycle associated protein 1b |
| chr1_+_38142354 | 1.58 |
ENSDART00000179352
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr14_-_31618243 | 1.58 |
ENSDART00000016592
|
mmgt1
|
membrane magnesium transporter 1 |
| chr23_+_13533885 | 1.58 |
ENSDART00000144386
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr11_+_19433936 | 1.58 |
ENSDART00000162081
|
prickle2b
|
prickle homolog 2b |
| chr10_-_3427589 | 1.57 |
ENSDART00000133452
ENSDART00000037183 |
tmed2
|
transmembrane p24 trafficking protein 2 |
| chr7_-_49800755 | 1.55 |
ENSDART00000180072
|
fjx1
|
four jointed box 1 |
| chr4_+_9011448 | 1.54 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr15_-_41689981 | 1.53 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr6_+_40992409 | 1.52 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr14_+_989733 | 1.51 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr13_+_37653851 | 1.51 |
ENSDART00000141988
ENSDART00000126902 ENSDART00000100352 |
phf3
|
PHD finger protein 3 |
| chr24_+_37800102 | 1.49 |
ENSDART00000187591
|
telo2
|
TEL2, telomere maintenance 2, homolog (S. cerevisiae) |
| chr22_+_30047245 | 1.47 |
ENSDART00000142857
ENSDART00000141247 ENSDART00000140015 ENSDART00000040538 |
add3a
|
adducin 3 (gamma) a |
| chr17_-_6955082 | 1.47 |
ENSDART00000109228
|
zbtb24
|
zinc finger and BTB domain containing 24 |
| chr2_-_42492445 | 1.47 |
ENSDART00000139929
|
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr23_-_16692312 | 1.47 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr5_+_1624359 | 1.47 |
ENSDART00000165431
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr5_-_29531948 | 1.46 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr25_-_25575241 | 1.45 |
ENSDART00000150636
|
hic1l
|
hypermethylated in cancer 1 like |
| chr8_+_18010978 | 1.45 |
ENSDART00000039887
ENSDART00000144532 |
ssbp3b
|
single stranded DNA binding protein 3b |
| chr24_-_25144441 | 1.44 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr14_+_3507326 | 1.44 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr16_+_35535375 | 1.44 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr14_-_24110251 | 1.44 |
ENSDART00000079226
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr17_+_23730089 | 1.43 |
ENSDART00000034913
|
zgc:91976
|
zgc:91976 |
| chr15_+_28410664 | 1.43 |
ENSDART00000132028
ENSDART00000057697 ENSDART00000057257 |
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr5_-_48680580 | 1.43 |
ENSDART00000031194
|
lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr6_+_27991943 | 1.43 |
ENSDART00000143974
ENSDART00000141354 ENSDART00000088914 ENSDART00000139367 |
amotl2a
|
angiomotin like 2a |
| chr6_-_39006449 | 1.42 |
ENSDART00000150885
|
vdrb
|
vitamin D receptor b |
| chr8_+_26868105 | 1.42 |
ENSDART00000005337
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr25_+_16194979 | 1.42 |
ENSDART00000185592
ENSDART00000158582 ENSDART00000161109 ENSDART00000139013 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr14_-_16754262 | 1.41 |
ENSDART00000001159
|
mgat4b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr17_-_6954719 | 1.39 |
ENSDART00000188180
|
zbtb24
|
zinc finger and BTB domain containing 24 |
| chr5_-_23118290 | 1.39 |
ENSDART00000132857
|
uprt
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr14_-_24110062 | 1.38 |
ENSDART00000177062
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr5_+_59030096 | 1.38 |
ENSDART00000179814
ENSDART00000110182 |
rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr9_-_28255029 | 1.37 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr12_+_36428052 | 1.35 |
ENSDART00000131300
|
unk
|
unkempt family zinc finger |
| chr14_-_1200854 | 1.35 |
ENSDART00000106672
|
arl9
|
ADP-ribosylation factor-like 9 |
| chr17_+_50261603 | 1.35 |
ENSDART00000154503
ENSDART00000154467 |
syncripl
|
synaptotagmin binding, cytoplasmic RNA interacting protein, like |
| chr1_+_14454663 | 1.34 |
ENSDART00000005067
|
rbpja
|
recombination signal binding protein for immunoglobulin kappa J region a |
| chr8_+_44623540 | 1.33 |
ENSDART00000141513
|
grk5l
|
G protein-coupled receptor kinase 5 like |
| chr11_+_31324335 | 1.33 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr16_+_19637384 | 1.33 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr16_+_33144306 | 1.32 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr11_+_6902946 | 1.31 |
ENSDART00000144006
|
crtc1b
|
CREB regulated transcription coactivator 1b |
| chr1_+_6226135 | 1.30 |
ENSDART00000108816
|
fastkd2
|
FAST kinase domains 2 |
| chr12_-_34852342 | 1.29 |
ENSDART00000152968
|
si:dkey-21c1.1
|
si:dkey-21c1.1 |
| chr15_-_41689684 | 1.28 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr24_-_38138307 | 1.27 |
ENSDART00000189178
|
crp7
|
C-reactive protein 7 |
| chr23_+_9230431 | 1.27 |
ENSDART00000187676
ENSDART00000125122 ENSDART00000190980 ENSDART00000141235 |
acss2
|
acyl-CoA synthetase short chain family member 2 |
| chr22_+_737211 | 1.26 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr24_-_34335265 | 1.26 |
ENSDART00000128690
|
agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr21_-_32036597 | 1.26 |
ENSDART00000114964
|
zgc:165573
|
zgc:165573 |
| chr18_-_39787040 | 1.25 |
ENSDART00000169916
|
dmxl2
|
Dmx-like 2 |
| chr18_+_6857071 | 1.25 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr23_+_35847538 | 1.25 |
ENSDART00000143935
|
rarga
|
retinoic acid receptor gamma a |
| chr3_-_20118342 | 1.25 |
ENSDART00000139902
|
selenow2a
|
selenoprotein W, 2a |
| chr1_-_36772147 | 1.24 |
ENSDART00000053369
|
prmt9
|
protein arginine methyltransferase 9 |
| chr7_-_18877109 | 1.24 |
ENSDART00000113593
|
mllt3
|
MLLT3, super elongation complex subunit |
| chr8_-_18010735 | 1.24 |
ENSDART00000125014
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr4_-_25181552 | 1.23 |
ENSDART00000066930
|
atp5f1c
|
ATP synthase F1 subunit gamma |
Network of associatons between targets according to the STRING database.
First level regulatory network of arnt2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.3 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.9 | 2.6 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.7 | 5.0 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.7 | 2.7 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.7 | 2.6 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.6 | 8.2 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.6 | 5.6 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.6 | 1.8 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.6 | 1.7 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 0.5 | 2.7 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.5 | 2.1 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.5 | 2.6 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.5 | 1.5 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.5 | 6.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.5 | 2.0 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.5 | 2.4 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.5 | 2.8 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.5 | 2.4 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.4 | 3.5 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.4 | 5.4 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.4 | 1.3 | GO:0097401 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.4 | 1.7 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.4 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.4 | 2.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.4 | 2.8 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.4 | 1.9 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.4 | 2.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 | 2.5 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.4 | 2.5 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.3 | 4.1 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.3 | 1.3 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.3 | 1.0 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.3 | 2.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.3 | 1.0 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.3 | 0.9 | GO:0046833 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.3 | 1.2 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.3 | 1.2 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.3 | 2.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.3 | 1.4 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.3 | 2.1 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.3 | 4.4 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.3 | 0.5 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.2 | 2.2 | GO:0045741 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.2 | 1.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 1.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 0.9 | GO:0021755 | eurydendroid cell differentiation(GO:0021755) |
| 0.2 | 1.6 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.2 | 0.7 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.2 | 0.7 | GO:0036314 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.2 | 1.1 | GO:0032196 | transposition(GO:0032196) |
| 0.2 | 0.9 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.2 | 2.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 1.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 0.6 | GO:0046048 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.2 | 1.3 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.2 | 1.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) anterior lateral line nerve development(GO:0048909) |
| 0.2 | 1.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.2 | 2.5 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.2 | 1.6 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.2 | 3.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.9 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 0.9 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.2 | 0.7 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.2 | 1.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 1.4 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.2 | 0.7 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 1.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 2.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.2 | 1.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 5.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 2.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 0.6 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.7 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 3.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 4.0 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.1 | 0.4 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 3.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.5 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 1.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.9 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 1.1 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 0.9 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.5 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.1 | 1.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 3.6 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 1.7 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 2.6 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.1 | 1.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.3 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 3.0 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 0.6 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.1 | 2.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.4 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 0.5 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 2.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.6 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 0.5 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.4 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 1.3 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 0.4 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.1 | 1.2 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 2.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 4.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 1.4 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 7.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 3.1 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 2.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.3 | GO:0019860 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.1 | 1.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.1 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.5 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 0.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 1.4 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 0.7 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 1.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 4.5 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 0.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 2.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.3 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 2.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.9 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 3.1 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 2.1 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 1.0 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.1 | GO:2000257 | complement activation, alternative pathway(GO:0006957) regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.9 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.0 | 1.7 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 1.6 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.2 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 1.6 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 1.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.0 | 2.1 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.0 | GO:0051784 | negative regulation of nuclear division(GO:0051784) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 2.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.6 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 3.1 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.2 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 1.4 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.9 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.9 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 2.6 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 1.2 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 2.6 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 1.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 5.2 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 1.9 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.9 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.9 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.2 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.9 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.2 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.0 | 1.6 | GO:0031929 | TOR signaling(GO:0031929) |
| 0.0 | 0.1 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.0 | 0.1 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.0 | 0.4 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 1.4 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 1.0 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.0 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
| 0.0 | 1.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.6 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.5 | 2.7 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.4 | 1.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.4 | 1.9 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.4 | 7.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.3 | 5.5 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.3 | 5.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 4.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 1.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 0.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 1.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.3 | 1.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.2 | 1.0 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.2 | 1.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 2.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 1.4 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.2 | 0.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 2.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 1.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 2.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 1.5 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.2 | 1.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 1.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 0.5 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 0.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 2.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.8 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.4 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.6 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 1.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 2.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 2.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.6 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 5.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 3.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 4.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.4 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.1 | 0.6 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.9 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 1.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 2.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.7 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 6.4 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 1.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 3.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.7 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 2.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.6 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.9 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 1.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 2.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 6.6 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 22.1 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 1.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0044224 | paranode region of axon(GO:0033270) juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 3.5 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.6 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.4 | GO:0019867 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 1.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.6 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.3 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 1.0 | 3.9 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.9 | 2.6 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.6 | 1.7 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.5 | 2.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.5 | 1.4 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.5 | 1.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 1.7 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.4 | 1.6 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.4 | 0.8 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.4 | 3.6 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.4 | 1.8 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.4 | 7.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.4 | 2.5 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.3 | 1.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 1.6 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.3 | 1.6 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.3 | 1.6 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.3 | 3.8 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 5.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 7.3 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.3 | 2.7 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.3 | 6.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.3 | 1.5 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.3 | 1.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.3 | 0.9 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.3 | 2.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.3 | 1.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.3 | 1.6 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.3 | 1.3 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.2 | 1.0 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.2 | 8.7 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.2 | 1.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 1.4 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.2 | 4.0 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.2 | 3.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 1.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 0.6 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 1.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.2 | 1.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 1.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 2.0 | GO:0005487 | nuclear export signal receptor activity(GO:0005049) nucleocytoplasmic transporter activity(GO:0005487) |
| 0.2 | 1.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 5.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 1.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 2.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 0.9 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 1.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 0.8 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.2 | 2.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.2 | 5.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 4.1 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.2 | 0.9 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 3.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 1.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 3.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 9.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 2.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 2.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 2.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 2.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 1.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 0.7 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 1.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 1.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 3.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 4.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 2.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.5 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 2.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 3.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 4.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.3 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.3 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 0.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.6 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.9 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.4 | GO:0001032 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 0.2 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.1 | 1.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 6.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 3.7 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0005334 | dopamine transmembrane transporter activity(GO:0005329) dopamine:sodium symporter activity(GO:0005330) norepinephrine transmembrane transporter activity(GO:0005333) norepinephrine:sodium symporter activity(GO:0005334) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 4.4 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 2.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 3.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 5.6 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 4.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 2.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.5 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 2.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 9.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 2.1 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.9 | GO:0000146 | microfilament motor activity(GO:0000146) actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 2.9 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 2.2 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 6.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 2.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 3.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 9.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 2.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 0.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 3.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 1.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 1.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 2.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 1.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 1.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 2.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 2.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 5.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 3.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.1 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 1.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.7 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.1 | 3.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 0.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 6.2 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 2.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.9 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.9 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 0.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |