Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
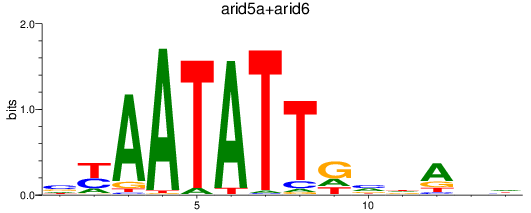
Results for arid5a+arid6
Z-value: 0.44
Transcription factors associated with arid5a+arid6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arid6
|
ENSDARG00000069988 | AT-rich interaction domain 6 |
|
arid5a
|
ENSDARG00000077120 | AT rich interactive domain 5A (MRF1-like) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arid6 | dr11_v1_chr21_+_11244068_11244068 | -0.47 | 4.8e-02 | Click! |
| arid5a | dr11_v1_chr8_+_52377516_52377516 | -0.04 | 8.7e-01 | Click! |
Activity profile of arid5a+arid6 motif
Sorted Z-values of arid5a+arid6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_25753972 | 1.22 |
ENSDART00000188417
|
AL929192.2
|
|
| chr3_+_39566999 | 0.87 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr22_+_25704430 | 0.78 |
ENSDART00000143776
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr20_+_54024559 | 0.74 |
ENSDART00000130767
|
si:dkey-241l7.4
|
si:dkey-241l7.4 |
| chr22_+_25715925 | 0.73 |
ENSDART00000150650
|
si:dkeyp-98a7.7
|
si:dkeyp-98a7.7 |
| chr22_+_25687525 | 0.70 |
ENSDART00000135717
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr22_+_25681911 | 0.69 |
ENSDART00000113381
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr22_+_25693295 | 0.69 |
ENSDART00000123888
ENSDART00000150783 |
si:dkeyp-98a7.4
si:dkeyp-98a7.3
|
si:dkeyp-98a7.4 si:dkeyp-98a7.3 |
| chr19_-_1948236 | 0.68 |
ENSDART00000163344
|
znrf2a
|
zinc and ring finger 2a |
| chr21_-_2299002 | 0.61 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr5_+_25074648 | 0.52 |
ENSDART00000188951
|
mrpl41
|
mitochondrial ribosomal protein L41 |
| chr10_+_44699734 | 0.51 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr5_-_2721686 | 0.50 |
ENSDART00000169404
|
hspa5
|
heat shock protein 5 |
| chr9_+_41040294 | 0.48 |
ENSDART00000000693
|
osgepl1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr4_-_8043839 | 0.48 |
ENSDART00000190047
ENSDART00000057567 |
si:ch211-240l19.5
|
si:ch211-240l19.5 |
| chr2_+_54389750 | 0.47 |
ENSDART00000189236
|
rab12
|
RAB12, member RAS oncogene family |
| chr22_-_557965 | 0.47 |
ENSDART00000001201
|
bysl
|
bystin-like |
| chr16_-_48400639 | 0.47 |
ENSDART00000159372
|
eif3ha
|
eukaryotic translation initiation factor 3, subunit H, a |
| chr24_+_7322116 | 0.45 |
ENSDART00000005804
|
XRCC2
|
X-ray repair cross complementing 2 |
| chr6_+_52235441 | 0.45 |
ENSDART00000056319
|
cox6c
|
cytochrome c oxidase subunit VIc |
| chr11_-_11331052 | 0.44 |
ENSDART00000081765
ENSDART00000160247 |
mrpl4
|
mitochondrial ribosomal protein L4 |
| chr3_+_24207243 | 0.44 |
ENSDART00000023454
ENSDART00000136400 |
adsl
|
adenylosuccinate lyase |
| chr1_+_16073887 | 0.44 |
ENSDART00000160270
|
tusc3
|
tumor suppressor candidate 3 |
| chr22_-_18491813 | 0.43 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr19_-_1947403 | 0.42 |
ENSDART00000113951
ENSDART00000151293 ENSDART00000134074 |
znrf2a
|
zinc and ring finger 2a |
| chr10_+_39091353 | 0.41 |
ENSDART00000125986
|
si:ch73-1a9.4
|
si:ch73-1a9.4 |
| chr10_-_17222083 | 0.41 |
ENSDART00000134059
|
depdc5
|
DEP domain containing 5 |
| chr20_+_36628059 | 0.41 |
ENSDART00000062898
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr6_+_13787855 | 0.39 |
ENSDART00000182899
|
tmem198b
|
transmembrane protein 198b |
| chr19_+_11978209 | 0.38 |
ENSDART00000111568
|
polr2k
|
polymerase (RNA) II (DNA directed) polypeptide K |
| chr18_+_19456648 | 0.38 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr19_+_816208 | 0.36 |
ENSDART00000093304
|
nrm
|
nurim |
| chr3_-_55104310 | 0.36 |
ENSDART00000101713
|
hbba1
|
hemoglobin, beta adult 1 |
| chr12_+_48220584 | 0.36 |
ENSDART00000164392
|
lrrc20
|
leucine rich repeat containing 20 |
| chr21_-_3422635 | 0.36 |
ENSDART00000150975
|
smad7
|
SMAD family member 7 |
| chr22_+_15343953 | 0.35 |
ENSDART00000045682
|
rrp36
|
ribosomal RNA processing 36 |
| chr3_+_31039923 | 0.35 |
ENSDART00000147706
|
cox6a2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr18_-_19456269 | 0.34 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr24_+_26328787 | 0.34 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr1_+_53714734 | 0.33 |
ENSDART00000114689
|
pus10
|
pseudouridylate synthase 10 |
| chr20_+_14114258 | 0.33 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr1_-_54063520 | 0.33 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr13_-_5064481 | 0.33 |
ENSDART00000091806
ENSDART00000032322 |
abcg2c
|
ATP-binding cassette, sub-family G (WHITE), member 2c |
| chr23_-_35483163 | 0.32 |
ENSDART00000138660
ENSDART00000113643 ENSDART00000189269 |
fbxo25
|
F-box protein 25 |
| chr11_-_438294 | 0.32 |
ENSDART00000040812
|
nuf2
|
NUF2, NDC80 kinetochore complex component, homolog |
| chr16_+_9762261 | 0.32 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr8_-_53535262 | 0.32 |
ENSDART00000167839
ENSDART00000157521 |
actr8
|
ARP8 actin related protein 8 homolog |
| chr25_-_1323623 | 0.31 |
ENSDART00000156532
ENSDART00000157163 ENSDART00000156062 ENSDART00000082447 ENSDART00000189175 |
calml4b
|
calmodulin-like 4b |
| chr1_-_53714885 | 0.30 |
ENSDART00000026409
|
cct4
|
chaperonin containing TCP1, subunit 4 (delta) |
| chr21_+_45502621 | 0.30 |
ENSDART00000166719
|
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr18_+_7594012 | 0.30 |
ENSDART00000062150
|
zgc:77752
|
zgc:77752 |
| chr11_-_574423 | 0.30 |
ENSDART00000176364
ENSDART00000130504 |
mkrn2
|
makorin, ring finger protein, 2 |
| chr7_-_12968689 | 0.29 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr16_-_46524842 | 0.29 |
ENSDART00000169477
|
tmem176l.1
|
transmembrane protein 176l.1 |
| chr22_-_2959005 | 0.29 |
ENSDART00000040701
|
ing5a
|
inhibitor of growth family, member 5a |
| chr19_-_41405187 | 0.29 |
ENSDART00000038038
ENSDART00000172350 |
sem1
|
SEM1, 26S proteasome complex subunit |
| chr24_-_9991153 | 0.29 |
ENSDART00000137794
ENSDART00000106252 ENSDART00000188309 ENSDART00000188266 ENSDART00000188660 ENSDART00000185713 ENSDART00000179773 |
zgc:152652
|
zgc:152652 |
| chr8_-_46321889 | 0.29 |
ENSDART00000075189
ENSDART00000122801 |
mtor
|
mechanistic target of rapamycin kinase |
| chr19_+_1465004 | 0.28 |
ENSDART00000159157
|
CABZ01073736.1
|
|
| chr24_-_35561672 | 0.28 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr25_-_774350 | 0.28 |
ENSDART00000166321
ENSDART00000160386 |
IRAK4
|
interleukin 1 receptor associated kinase 4 |
| chr11_-_44906703 | 0.28 |
ENSDART00000180591
|
zgc:171772
|
zgc:171772 |
| chr16_+_52847079 | 0.28 |
ENSDART00000163151
|
cep72
|
centrosomal protein 72 |
| chr23_+_19670085 | 0.28 |
ENSDART00000031872
|
kctd6b
|
potassium channel tetramerization domain containing 6b |
| chr24_-_36271352 | 0.27 |
ENSDART00000153682
ENSDART00000155892 |
si:ch211-40k21.5
|
si:ch211-40k21.5 |
| chr19_-_1363759 | 0.27 |
ENSDART00000166088
|
zgc:63863
|
zgc:63863 |
| chr12_-_47648538 | 0.26 |
ENSDART00000108477
|
fh
|
fumarate hydratase |
| chr17_-_4395373 | 0.26 |
ENSDART00000015923
|
klhl10a
|
kelch-like family member 10a |
| chr21_-_8003782 | 0.26 |
ENSDART00000126792
|
si:dkey-163m14.7
|
si:dkey-163m14.7 |
| chr9_-_18911608 | 0.26 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr5_+_37837245 | 0.26 |
ENSDART00000171617
|
epd
|
ependymin |
| chr4_-_72609118 | 0.26 |
ENSDART00000174077
|
si:cabz01054396.2
|
si:cabz01054396.2 |
| chr4_+_72723304 | 0.26 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr21_+_45502773 | 0.25 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr25_+_34845115 | 0.25 |
ENSDART00000061996
|
tmem231
|
transmembrane protein 231 |
| chr5_+_4366431 | 0.25 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr15_+_32387063 | 0.25 |
ENSDART00000154210
ENSDART00000156525 |
si:ch211-162k9.5
|
si:ch211-162k9.5 |
| chr11_-_41853874 | 0.25 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr13_-_12021566 | 0.25 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr24_-_7632187 | 0.25 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr23_+_45512825 | 0.25 |
ENSDART00000064846
|
prelid1b
|
PRELI domain containing 1b |
| chr22_-_15562933 | 0.24 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr5_+_22177033 | 0.24 |
ENSDART00000131223
|
fdx1b
|
ferredoxin 1b |
| chr2_-_3611960 | 0.24 |
ENSDART00000184579
|
pter
|
phosphotriesterase related |
| chr21_+_10577527 | 0.24 |
ENSDART00000165070
ENSDART00000127963 |
ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr25_+_32755485 | 0.24 |
ENSDART00000162188
|
etfa
|
electron-transfer-flavoprotein, alpha polypeptide |
| chr17_-_31579715 | 0.24 |
ENSDART00000110167
ENSDART00000191092 |
rpap1
|
RNA polymerase II associated protein 1 |
| chr12_+_34891529 | 0.24 |
ENSDART00000015643
|
tbcc
|
tubulin folding cofactor C |
| chr10_+_45089820 | 0.23 |
ENSDART00000175481
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr16_-_32128345 | 0.23 |
ENSDART00000102100
ENSDART00000137699 |
cnot3a
|
CCR4-NOT transcription complex, subunit 3a |
| chr19_-_17210760 | 0.23 |
ENSDART00000007906
|
stmn1a
|
stathmin 1a |
| chr17_+_37227936 | 0.23 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr10_+_15777258 | 0.23 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr23_+_43638982 | 0.23 |
ENSDART00000168646
|
slc10a7
|
solute carrier family 10, member 7 |
| chr10_+_31222433 | 0.23 |
ENSDART00000185080
|
tmem218
|
transmembrane protein 218 |
| chr8_+_34434345 | 0.23 |
ENSDART00000190246
ENSDART00000189447 ENSDART00000185557 ENSDART00000189230 |
zgc:174461
|
zgc:174461 |
| chr8_+_41229233 | 0.23 |
ENSDART00000131135
|
zgc:152830
|
zgc:152830 |
| chr24_-_26328721 | 0.22 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr14_-_12822 | 0.22 |
ENSDART00000180650
ENSDART00000188819 |
msx1a
|
muscle segment homeobox 1a |
| chr21_+_20903244 | 0.22 |
ENSDART00000186193
|
c7b
|
complement component 7b |
| chr16_-_46525135 | 0.22 |
ENSDART00000169940
ENSDART00000163180 |
tmem176l.1
|
transmembrane protein 176l.1 |
| chr2_+_9990491 | 0.22 |
ENSDART00000011906
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr25_-_6223567 | 0.22 |
ENSDART00000067512
|
psma4
|
proteasome subunit alpha 4 |
| chr6_-_33878665 | 0.22 |
ENSDART00000129916
|
tmem69
|
transmembrane protein 69 |
| chr22_-_11623063 | 0.22 |
ENSDART00000145653
|
gcga
|
glucagon a |
| chr22_-_9889413 | 0.22 |
ENSDART00000081450
|
si:dkey-253d23.5
|
si:dkey-253d23.5 |
| chr7_+_12931396 | 0.21 |
ENSDART00000184664
|
fth1a
|
ferritin, heavy polypeptide 1a |
| chr12_-_35977702 | 0.21 |
ENSDART00000165935
|
nploc4
|
NPL4 homolog, ubiquitin recognition factor |
| chr11_-_38083397 | 0.21 |
ENSDART00000086516
ENSDART00000184033 |
klhdc8a
|
kelch domain containing 8A |
| chr22_-_17611742 | 0.21 |
ENSDART00000144031
|
gpx4a
|
glutathione peroxidase 4a |
| chr23_-_45407631 | 0.21 |
ENSDART00000148484
ENSDART00000150186 |
zgc:101853
|
zgc:101853 |
| chr20_-_26822522 | 0.21 |
ENSDART00000146326
ENSDART00000046764 ENSDART00000103234 ENSDART00000143267 |
gmds
|
GDP-mannose 4,6-dehydratase |
| chr20_-_48061351 | 0.21 |
ENSDART00000164962
|
prep
|
prolyl endopeptidase |
| chr22_+_31039091 | 0.20 |
ENSDART00000060005
|
rpl32
|
ribosomal protein L32 |
| chr3_-_62403550 | 0.20 |
ENSDART00000055055
|
sox8b
|
SRY (sex determining region Y)-box 8b |
| chr16_+_2565433 | 0.20 |
ENSDART00000188014
ENSDART00000171378 |
preb
|
prolactin regulatory element binding |
| chr24_-_36270855 | 0.20 |
ENSDART00000154858
|
si:ch211-40k21.5
|
si:ch211-40k21.5 |
| chr12_-_46959990 | 0.20 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr19_+_47405867 | 0.19 |
ENSDART00000041114
|
psmb2
|
proteasome subunit beta 2 |
| chr7_+_21275152 | 0.19 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr7_+_6879534 | 0.19 |
ENSDART00000157731
|
zgc:175248
|
zgc:175248 |
| chr22_+_1421212 | 0.19 |
ENSDART00000161813
|
zgc:101130
|
zgc:101130 |
| chr3_-_61205711 | 0.19 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr8_-_22508055 | 0.19 |
ENSDART00000101616
|
si:ch211-261n11.5
|
si:ch211-261n11.5 |
| chr7_-_30925798 | 0.19 |
ENSDART00000149303
|
sord
|
sorbitol dehydrogenase |
| chr7_-_30926030 | 0.19 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr8_+_49117518 | 0.19 |
ENSDART00000079631
|
rad21l1
|
RAD21 cohesin complex component like 1 |
| chr18_+_44630415 | 0.19 |
ENSDART00000098540
|
bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr23_+_19655301 | 0.19 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr24_+_3328354 | 0.19 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr3_+_62356578 | 0.18 |
ENSDART00000157030
|
iqck
|
IQ motif containing K |
| chr7_-_18168493 | 0.18 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr21_-_15929041 | 0.18 |
ENSDART00000080693
|
lhx5
|
LIM homeobox 5 |
| chr21_+_25533531 | 0.18 |
ENSDART00000134052
|
nlrc3l1
|
NLR family, CARD domain containing 3-like 1 |
| chr11_-_3954691 | 0.18 |
ENSDART00000182041
|
pbrm1
|
polybromo 1 |
| chr18_+_20468157 | 0.18 |
ENSDART00000100665
ENSDART00000147867 ENSDART00000060302 ENSDART00000180370 |
ddb2
|
damage-specific DNA binding protein 2 |
| chr9_-_42696408 | 0.18 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr14_+_9581896 | 0.18 |
ENSDART00000114563
|
tmem129
|
transmembrane protein 129, E3 ubiquitin protein ligase |
| chr14_-_4121052 | 0.18 |
ENSDART00000167074
|
irf2
|
interferon regulatory factor 2 |
| chr19_+_33139164 | 0.18 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr11_+_6881001 | 0.17 |
ENSDART00000170331
|
klhl26
|
kelch-like family member 26 |
| chr10_-_41940302 | 0.17 |
ENSDART00000033121
|
morn3
|
MORN repeat containing 3 |
| chr2_-_20574193 | 0.17 |
ENSDART00000190448
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr8_+_7975745 | 0.17 |
ENSDART00000137920
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr25_+_7571920 | 0.17 |
ENSDART00000042928
|
fuk
|
fucokinase |
| chr14_+_5383060 | 0.17 |
ENSDART00000187825
|
lbx2
|
ladybird homeobox 2 |
| chr23_+_38806861 | 0.17 |
ENSDART00000086879
|
zfp64
|
zinc finger protein 64 homolog (mouse) |
| chr3_-_13461056 | 0.17 |
ENSDART00000137678
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr13_-_22907260 | 0.17 |
ENSDART00000143097
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr16_-_46393154 | 0.17 |
ENSDART00000132154
|
si:ch73-59c19.1
|
si:ch73-59c19.1 |
| chr15_-_23647078 | 0.17 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr3_-_13461361 | 0.16 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr10_+_15777064 | 0.16 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr4_+_391297 | 0.16 |
ENSDART00000030215
|
rpl18a
|
ribosomal protein L18a |
| chr12_+_46696867 | 0.16 |
ENSDART00000152928
ENSDART00000153445 ENSDART00000123357 ENSDART00000152880 ENSDART00000123834 ENSDART00000174765 ENSDART00000189923 |
exoc7
|
exocyst complex component 7 |
| chr14_+_8475007 | 0.16 |
ENSDART00000148210
|
dnajc4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr14_+_31865099 | 0.16 |
ENSDART00000189124
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr7_+_62248514 | 0.16 |
ENSDART00000025308
|
tbc1d19
|
TBC1 domain family, member 19 |
| chr16_+_40576679 | 0.15 |
ENSDART00000169412
ENSDART00000193464 |
ccne2
|
cyclin E2 |
| chr20_-_14114078 | 0.15 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr11_-_287670 | 0.15 |
ENSDART00000035737
|
slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr20_+_5985329 | 0.15 |
ENSDART00000165489
|
cep128
|
centrosomal protein 128 |
| chr22_+_7439186 | 0.15 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr24_+_23716918 | 0.15 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr20_+_30578967 | 0.15 |
ENSDART00000010494
|
fgfr1op
|
FGFR1 oncogene partner |
| chr25_-_26753196 | 0.15 |
ENSDART00000155698
|
usp3
|
ubiquitin specific peptidase 3 |
| chr25_+_37209619 | 0.15 |
ENSDART00000112192
|
si:dkey-234i14.3
|
si:dkey-234i14.3 |
| chr25_-_8138122 | 0.15 |
ENSDART00000104659
|
sergef
|
secretion regulating guanine nucleotide exchange factor |
| chr18_+_25752592 | 0.15 |
ENSDART00000111767
|
si:ch211-39k3.2
|
si:ch211-39k3.2 |
| chr16_-_12173554 | 0.15 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr11_+_30647545 | 0.14 |
ENSDART00000114792
|
gb:eh507706
|
expressed sequence EH507706 |
| chr1_+_21731382 | 0.14 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr2_-_3045861 | 0.14 |
ENSDART00000105818
ENSDART00000187575 |
guk1a
|
guanylate kinase 1a |
| chr24_+_39695688 | 0.14 |
ENSDART00000109747
|
zgc:153659
|
zgc:153659 |
| chr2_-_7605121 | 0.14 |
ENSDART00000182099
|
CABZ01021599.1
|
|
| chr5_-_31716713 | 0.14 |
ENSDART00000131443
|
dpm2
|
dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit |
| chr19_+_43119014 | 0.14 |
ENSDART00000023156
|
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr9_+_37329036 | 0.14 |
ENSDART00000131756
|
slc15a2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chr23_+_383782 | 0.14 |
ENSDART00000055148
|
zgc:101663
|
zgc:101663 |
| chr4_-_4834617 | 0.14 |
ENSDART00000141539
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr13_-_24018916 | 0.14 |
ENSDART00000026189
|
cog2
|
component of oligomeric golgi complex 2 |
| chr10_-_39130839 | 0.14 |
ENSDART00000061274
ENSDART00000148648 |
rps25
|
ribosomal protein S25 |
| chr23_+_17220986 | 0.14 |
ENSDART00000054761
|
nol4lb
|
nucleolar protein 4-like b |
| chr25_+_36325793 | 0.14 |
ENSDART00000186973
|
HIST1H4E
|
zgc:165555 |
| chr15_-_35252522 | 0.14 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr18_+_36769758 | 0.14 |
ENSDART00000180375
ENSDART00000136463 ENSDART00000133487 ENSDART00000130206 |
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr4_-_9637398 | 0.14 |
ENSDART00000150253
|
dclre1c
|
DNA cross-link repair 1C, PSO2 homolog (S. cerevisiae) |
| chr24_-_22756508 | 0.13 |
ENSDART00000035409
ENSDART00000146247 |
zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr25_-_17494364 | 0.13 |
ENSDART00000154134
|
lrrc29
|
leucine rich repeat containing 29 |
| chr7_+_48806420 | 0.13 |
ENSDART00000083431
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr11_-_6989598 | 0.13 |
ENSDART00000102493
ENSDART00000173242 ENSDART00000172896 |
zgc:173548
|
zgc:173548 |
| chr24_-_31306724 | 0.13 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr19_-_3973567 | 0.13 |
ENSDART00000167580
|
map7d1b
|
MAP7 domain containing 1b |
| chr1_-_49530615 | 0.13 |
ENSDART00000113315
|
si:ch211-281g13.5
|
si:ch211-281g13.5 |
| chr14_+_45566074 | 0.13 |
ENSDART00000133477
ENSDART00000185899 |
zgc:92249
|
zgc:92249 |
| chr9_+_13229585 | 0.13 |
ENSDART00000154879
ENSDART00000141705 |
carf
|
calcium responsive transcription factor |
| chr19_-_32600823 | 0.13 |
ENSDART00000134149
ENSDART00000187858 |
zgc:91944
|
zgc:91944 |
| chr7_+_15659280 | 0.13 |
ENSDART00000173414
|
mef2ab
|
myocyte enhancer factor 2ab |
| chr25_+_35133404 | 0.13 |
ENSDART00000188505
|
hist1h2a2
|
histone cluster 1 H2A family member 2 |
| chr25_+_10458990 | 0.13 |
ENSDART00000130354
ENSDART00000044738 |
ric8a
|
RIC8 guanine nucleotide exchange factor A |
Network of associatons between targets according to the STRING database.
First level regulatory network of arid5a+arid6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.4 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.1 | 0.5 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.3 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.2 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.4 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.4 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.0 | 0.1 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.0 | 0.4 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.0 | 0.4 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.9 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.4 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.3 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.3 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.2 | GO:0034650 | glucocorticoid biosynthetic process(GO:0006704) cortisol metabolic process(GO:0034650) |
| 0.0 | 0.2 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.0 | 0.2 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.5 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.1 | GO:0090183 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0032602 | chemokine production(GO:0032602) |
| 0.0 | 0.1 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.0 | 0.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.0 | 0.1 | GO:0035739 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.1 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.2 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.0 | 0.1 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.2 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.0 | 0.3 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.0 | GO:0003097 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.4 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0043247 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.0 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.2 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.0 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.0 | 0.0 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.0 | 0.1 | GO:2000316 | regulation of T-helper 17 type immune response(GO:2000316) negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) regulation of T-helper 17 cell lineage commitment(GO:2000328) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.0 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.2 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0030892 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.4 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.1 | 0.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.4 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.0 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.2 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.4 | GO:0019825 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 3.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 2.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | PID REELIN PATHWAY | Reelin signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.6 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |