Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
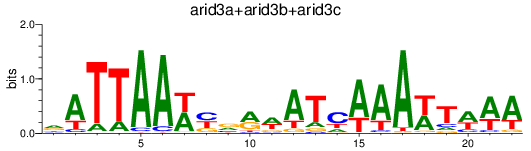
Results for arid3a+arid3b+arid3c
Z-value: 1.39
Transcription factors associated with arid3a+arid3b+arid3c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arid3c
|
ENSDARG00000067729 | AT rich interactive domain 3C (BRIGHT-like) |
|
arid3a
|
ENSDARG00000070843 | AT rich interactive domain 3A (BRIGHT-like) |
|
arid3b
|
ENSDARG00000104034 | AT rich interactive domain 3B (BRIGHT-like) |
|
arid3c
|
ENSDARG00000114447 | AT rich interactive domain 3C (BRIGHT-like) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arid3b | dr11_v1_chr25_-_29074064_29074064 | 0.89 | 6.5e-07 | Click! |
| arid3c | dr11_v1_chr5_+_41322783_41322783 | -0.77 | 2.0e-04 | Click! |
| arid3a | dr11_v1_chr11_+_5681762_5681762 | 0.71 | 8.8e-04 | Click! |
Activity profile of arid3a+arid3b+arid3c motif
Sorted Z-values of arid3a+arid3b+arid3c motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_45031398 | 7.77 |
ENSDART00000160536
|
gnsb
|
glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID), b |
| chr21_-_42831033 | 7.51 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr17_-_1174960 | 5.03 |
ENSDART00000161202
|
dnajc17
|
DnaJ (Hsp40) homolog, subfamily C, member 17 |
| chr17_-_4245902 | 4.76 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr14_+_46342882 | 4.31 |
ENSDART00000193707
ENSDART00000060577 |
tmem33
|
transmembrane protein 33 |
| chr15_-_31109760 | 3.47 |
ENSDART00000154254
|
ksr1b
|
kinase suppressor of ras 1b |
| chr5_+_69650148 | 3.41 |
ENSDART00000097244
|
gtf2h3
|
general transcription factor IIH, polypeptide 3 |
| chr8_-_29930821 | 3.40 |
ENSDART00000125173
|
ercc6l2
|
excision repair cross-complementation group 6-like 2 |
| chr12_+_1609563 | 3.03 |
ENSDART00000163559
|
SLC39A11
|
solute carrier family 39 member 11 |
| chr21_+_43172506 | 2.98 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr15_+_38308421 | 2.90 |
ENSDART00000129941
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr12_-_48188928 | 2.89 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr23_+_2666944 | 2.82 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr7_-_8712148 | 2.77 |
ENSDART00000065488
|
tex261
|
testis expressed 261 |
| chr7_+_9189547 | 2.76 |
ENSDART00000169783
|
pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr12_-_13650344 | 2.76 |
ENSDART00000124364
ENSDART00000124638 ENSDART00000171929 |
stat5b
|
signal transducer and activator of transcription 5b |
| chr4_-_5775507 | 2.69 |
ENSDART00000021753
|
ccnc
|
cyclin C |
| chr6_-_8360918 | 2.62 |
ENSDART00000004716
|
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr17_-_7060477 | 2.60 |
ENSDART00000081797
|
sash1b
|
SAM and SH3 domain containing 1b |
| chr7_+_36467315 | 2.52 |
ENSDART00000138893
|
aktip
|
akt interacting protein |
| chr10_+_16911951 | 2.49 |
ENSDART00000164933
|
UNC13B (1 of many)
|
unc-13 homolog B |
| chr18_+_11858397 | 2.48 |
ENSDART00000133762
|
tmtc2b
|
transmembrane and tetratricopeptide repeat containing 2b |
| chr5_+_44895949 | 2.48 |
ENSDART00000193822
|
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr3_+_7808459 | 2.41 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr21_-_20322816 | 2.40 |
ENSDART00000135472
|
zgc:86764
|
zgc:86764 |
| chr19_+_4139065 | 2.31 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr14_+_30340251 | 2.26 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr5_-_42180205 | 2.21 |
ENSDART00000145247
|
fam222ba
|
family with sequence similarity 222, member Ba |
| chr5_+_22579975 | 2.20 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr18_+_39416357 | 2.09 |
ENSDART00000183174
ENSDART00000127955 ENSDART00000171303 |
lysmd2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr12_-_18961289 | 2.09 |
ENSDART00000168405
|
ep300a
|
E1A binding protein p300 a |
| chr20_+_35208020 | 2.05 |
ENSDART00000153315
ENSDART00000045135 |
fbxo16
|
F-box protein 16 |
| chr1_-_52790724 | 2.02 |
ENSDART00000139577
ENSDART00000100937 |
patl1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr9_-_27398369 | 1.97 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr19_+_25971000 | 1.95 |
ENSDART00000089836
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr14_+_16287968 | 1.92 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr7_+_57088920 | 1.86 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr18_-_48296793 | 1.85 |
ENSDART00000032184
ENSDART00000193076 |
CABZ01069595.1
|
|
| chr24_-_15208414 | 1.83 |
ENSDART00000145978
|
rttn
|
rotatin |
| chr16_-_42461263 | 1.80 |
ENSDART00000109259
|
smarcc1a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1a |
| chr17_-_29213710 | 1.79 |
ENSDART00000076481
|
ehd4
|
EH-domain containing 4 |
| chr4_-_25911609 | 1.78 |
ENSDART00000186958
ENSDART00000175487 |
fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr2_-_57916317 | 1.75 |
ENSDART00000183930
|
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
| chr5_+_5398966 | 1.73 |
ENSDART00000139553
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr17_+_16046314 | 1.71 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr15_+_20530649 | 1.70 |
ENSDART00000186312
|
tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr7_-_51749683 | 1.70 |
ENSDART00000083190
|
hdac8
|
histone deacetylase 8 |
| chr24_+_37484661 | 1.70 |
ENSDART00000165125
ENSDART00000109221 |
wdr90
|
WD repeat domain 90 |
| chr22_+_2751887 | 1.63 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr23_-_24343363 | 1.59 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr17_+_22311413 | 1.58 |
ENSDART00000151929
|
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr2_+_25657958 | 1.58 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr7_-_51749895 | 1.58 |
ENSDART00000175523
ENSDART00000189639 |
hdac8
|
histone deacetylase 8 |
| chr3_-_34136368 | 1.54 |
ENSDART00000136900
ENSDART00000186125 |
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr14_+_35428152 | 1.54 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr14_-_16082806 | 1.54 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr22_-_10752471 | 1.51 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr4_-_13931508 | 1.50 |
ENSDART00000067174
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr24_-_32173754 | 1.49 |
ENSDART00000048633
|
trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr5_+_27404946 | 1.48 |
ENSDART00000121886
ENSDART00000005025 |
hdr
|
hematopoietic death receptor |
| chr19_-_7406933 | 1.48 |
ENSDART00000151137
|
oxr1b
|
oxidation resistance 1b |
| chr8_+_3820134 | 1.48 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr22_+_30047245 | 1.47 |
ENSDART00000142857
ENSDART00000141247 ENSDART00000140015 ENSDART00000040538 |
add3a
|
adducin 3 (gamma) a |
| chr11_-_2504750 | 1.45 |
ENSDART00000173149
|
dgkab
|
diacylglycerol kinase, alpha b |
| chr5_-_25576462 | 1.45 |
ENSDART00000165147
|
si:dkey-229d2.4
|
si:dkey-229d2.4 |
| chr12_+_38774860 | 1.43 |
ENSDART00000130371
|
kif19
|
kinesin family member 19 |
| chr11_-_43200994 | 1.42 |
ENSDART00000164700
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr11_+_2687395 | 1.42 |
ENSDART00000082510
|
b3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 |
| chr19_-_46018152 | 1.40 |
ENSDART00000159206
|
krit1
|
KRIT1, ankyrin repeat containing |
| chr2_-_10877765 | 1.39 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr5_-_67349916 | 1.37 |
ENSDART00000144092
|
mlxip
|
MLX interacting protein |
| chr4_-_13931293 | 1.37 |
ENSDART00000067172
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr10_+_10313024 | 1.37 |
ENSDART00000142895
ENSDART00000129952 |
urm1
|
ubiquitin related modifier 1 |
| chr4_+_17671164 | 1.34 |
ENSDART00000040445
|
gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr9_+_33158191 | 1.32 |
ENSDART00000180786
|
dopey2
|
dopey family member 2 |
| chr13_+_24671481 | 1.31 |
ENSDART00000001678
|
adam8a
|
ADAM metallopeptidase domain 8a |
| chr4_+_25912308 | 1.31 |
ENSDART00000167845
ENSDART00000136927 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr19_-_34063567 | 1.30 |
ENSDART00000157815
ENSDART00000183907 |
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr20_+_46741074 | 1.30 |
ENSDART00000145294
|
si:ch211-57i17.1
|
si:ch211-57i17.1 |
| chr13_+_25199849 | 1.29 |
ENSDART00000139209
ENSDART00000130876 |
ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr3_-_25377163 | 1.29 |
ENSDART00000055490
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr10_-_16868211 | 1.28 |
ENSDART00000171755
|
stoml2
|
stomatin (EPB72)-like 2 |
| chr19_-_11208782 | 1.25 |
ENSDART00000044426
ENSDART00000189754 |
si:dkey-240h12.4
|
si:dkey-240h12.4 |
| chr8_+_28358161 | 1.24 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr14_+_45028062 | 1.24 |
ENSDART00000184717
ENSDART00000185481 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr5_+_6672870 | 1.23 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr3_+_60828813 | 1.23 |
ENSDART00000128260
|
CABZ01087514.1
|
|
| chr16_+_22345513 | 1.21 |
ENSDART00000078000
|
zgc:123238
|
zgc:123238 |
| chr2_+_34967022 | 1.21 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr19_-_8798178 | 1.21 |
ENSDART00000188232
|
cers2a
|
ceramide synthase 2a |
| chr10_-_14943281 | 1.21 |
ENSDART00000143608
|
smad2
|
SMAD family member 2 |
| chr16_+_26747766 | 1.21 |
ENSDART00000183257
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr4_+_13931578 | 1.20 |
ENSDART00000142466
|
pphln1
|
periphilin 1 |
| chr16_-_4610255 | 1.20 |
ENSDART00000081852
ENSDART00000123253 ENSDART00000127554 ENSDART00000029485 |
arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr13_+_25200105 | 1.20 |
ENSDART00000039640
|
ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr19_+_32401278 | 1.19 |
ENSDART00000184353
|
atxn1a
|
ataxin 1a |
| chr9_+_33220342 | 1.19 |
ENSDART00000100893
ENSDART00000113451 |
si:ch211-125e6.13
|
si:ch211-125e6.13 |
| chr3_-_5228841 | 1.18 |
ENSDART00000092373
ENSDART00000182438 |
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr11_-_26832685 | 1.13 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr1_-_29061285 | 1.12 |
ENSDART00000053933
ENSDART00000142350 ENSDART00000192615 |
gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr23_-_20051369 | 1.11 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr12_+_2995887 | 1.11 |
ENSDART00000189499
ENSDART00000182597 ENSDART00000184918 ENSDART00000182292 |
lrrc45
|
leucine rich repeat containing 45 |
| chr10_+_20392083 | 1.10 |
ENSDART00000166867
|
r3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr1_+_22654875 | 1.09 |
ENSDART00000019698
ENSDART00000161874 |
anxa5b
|
annexin A5b |
| chr7_-_25133783 | 1.09 |
ENSDART00000173781
ENSDART00000121943 ENSDART00000077219 |
badb
|
BCL2 associated agonist of cell death b |
| chr2_-_57837838 | 1.08 |
ENSDART00000010699
|
sf3a2
|
splicing factor 3a, subunit 2 |
| chr18_+_6641542 | 1.07 |
ENSDART00000160379
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr11_-_25461336 | 1.06 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr2_-_39558643 | 1.05 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr14_-_7045009 | 1.05 |
ENSDART00000112082
|
rufy1
|
RUN and FYVE domain containing 1 |
| chr15_-_34408777 | 1.05 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr14_+_30413758 | 1.04 |
ENSDART00000092953
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr16_+_1100559 | 1.04 |
ENSDART00000092657
|
adamts16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16 |
| chr21_-_11970199 | 1.04 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr20_+_35247410 | 1.04 |
ENSDART00000153098
ENSDART00000182470 |
fbxo16
|
F-box protein 16 |
| chr1_-_51038885 | 1.03 |
ENSDART00000035150
|
spast
|
spastin |
| chr15_+_47362728 | 1.03 |
ENSDART00000180712
|
CABZ01099833.1
|
|
| chr3_+_22036113 | 1.02 |
ENSDART00000132190
|
cdc27
|
cell division cycle 27 |
| chr1_+_35956435 | 1.02 |
ENSDART00000085021
ENSDART00000148505 |
mmaa
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr9_+_34380299 | 1.01 |
ENSDART00000131705
|
lamp1
|
lysosomal-associated membrane protein 1 |
| chr10_+_15967643 | 1.00 |
ENSDART00000136709
|
apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr21_-_3606539 | 1.00 |
ENSDART00000062418
|
dym
|
dymeclin |
| chr23_-_37291793 | 1.00 |
ENSDART00000083281
ENSDART00000187108 |
mul1b
|
mitochondrial E3 ubiquitin protein ligase 1b |
| chr5_-_67365750 | 0.98 |
ENSDART00000062359
|
unga
|
uracil DNA glycosylase a |
| chr15_-_33807758 | 0.98 |
ENSDART00000158445
|
pds5b
|
PDS5 cohesin associated factor B |
| chr22_+_12366516 | 0.98 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr12_-_17593030 | 0.97 |
ENSDART00000168758
|
usp42
|
ubiquitin specific peptidase 42 |
| chr6_-_33871055 | 0.95 |
ENSDART00000115409
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr7_+_48297842 | 0.94 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr24_-_20152965 | 0.93 |
ENSDART00000147004
|
oxsr1b
|
oxidative stress responsive 1b |
| chr14_-_36763302 | 0.93 |
ENSDART00000074786
|
ctso
|
cathepsin O |
| chr4_+_13931733 | 0.93 |
ENSDART00000141742
ENSDART00000067175 |
pphln1
|
periphilin 1 |
| chr13_-_17943135 | 0.91 |
ENSDART00000176027
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr1_+_14020445 | 0.91 |
ENSDART00000079716
|
hpf1
|
histone PARylation factor 1 |
| chr12_-_33659328 | 0.91 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr16_+_32014552 | 0.91 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr5_-_67365333 | 0.87 |
ENSDART00000133438
|
unga
|
uracil DNA glycosylase a |
| chr23_+_32101202 | 0.85 |
ENSDART00000000992
|
zgc:56699
|
zgc:56699 |
| chr25_-_13549577 | 0.85 |
ENSDART00000166772
|
ano10b
|
anoctamin 10b |
| chr3_+_42999277 | 0.85 |
ENSDART00000163595
|
ints1
|
integrator complex subunit 1 |
| chr8_+_9715010 | 0.85 |
ENSDART00000139414
|
gripap1
|
GRIP1 associated protein 1 |
| chr22_-_3275888 | 0.84 |
ENSDART00000164743
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr3_+_15893039 | 0.83 |
ENSDART00000055780
|
jpt2
|
Jupiter microtubule associated homolog 2 |
| chr18_-_16885362 | 0.83 |
ENSDART00000132778
|
swap70b
|
switching B cell complex subunit SWAP70b |
| chr6_-_3998199 | 0.82 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr25_-_21031007 | 0.81 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr13_+_18371208 | 0.80 |
ENSDART00000138172
|
ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr2_-_50272077 | 0.79 |
ENSDART00000127623
|
cul1a
|
cullin 1a |
| chr19_-_24267823 | 0.79 |
ENSDART00000132430
|
s100v2
|
S100 calcium binding protein V2 |
| chr23_-_32100106 | 0.78 |
ENSDART00000044658
|
letmd1
|
LETM1 domain containing 1 |
| chr4_-_5019113 | 0.78 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr9_+_2574122 | 0.78 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr7_-_7692723 | 0.76 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr24_-_36187094 | 0.76 |
ENSDART00000111891
|
esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr5_-_33039670 | 0.76 |
ENSDART00000141361
|
glipr2
|
GLI pathogenesis-related 2 |
| chr6_+_40951227 | 0.76 |
ENSDART00000156660
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr3_-_52899394 | 0.75 |
ENSDART00000128223
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr11_+_31323746 | 0.75 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr3_+_46271911 | 0.74 |
ENSDART00000186557
ENSDART00000113531 |
mkl2b
|
MKL/myocardin-like 2b |
| chr7_-_7692992 | 0.74 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr5_+_66250856 | 0.74 |
ENSDART00000132789
|
secisbp2
|
SECIS binding protein 2 |
| chr9_+_38502524 | 0.74 |
ENSDART00000087229
|
lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr14_+_3507326 | 0.74 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr2_-_22530969 | 0.74 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr10_-_31104983 | 0.74 |
ENSDART00000186941
|
pknox2
|
pbx/knotted 1 homeobox 2 |
| chr6_-_3978919 | 0.73 |
ENSDART00000167753
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr20_+_54383838 | 0.72 |
ENSDART00000157737
|
lrfn5b
|
leucine rich repeat and fibronectin type III domain containing 5b |
| chr20_+_22799641 | 0.72 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr7_+_1530024 | 0.71 |
ENSDART00000163082
|
tox4b
|
TOX high mobility group box family member 4 b |
| chr23_+_4777348 | 0.71 |
ENSDART00000171971
|
vgll4a
|
vestigial-like family member 4a |
| chr12_-_17199381 | 0.71 |
ENSDART00000193292
|
lipf
|
lipase, gastric |
| chr13_+_5978809 | 0.70 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr3_+_17634952 | 0.70 |
ENSDART00000191833
|
rab5c
|
RAB5C, member RAS oncogene family |
| chr7_+_16047863 | 0.69 |
ENSDART00000188750
|
immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr6_-_16394528 | 0.68 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr20_-_27190393 | 0.68 |
ENSDART00000149024
|
btbd7
|
BTB (POZ) domain containing 7 |
| chr6_-_55309190 | 0.67 |
ENSDART00000162117
|
ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr6_-_57485411 | 0.67 |
ENSDART00000011663
|
snrpb
|
small nuclear ribonucleoprotein polypeptides B and B1 |
| chr15_+_39096736 | 0.67 |
ENSDART00000129511
ENSDART00000014877 |
robo2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr6_+_26948093 | 0.66 |
ENSDART00000153595
|
farp2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr13_+_31550185 | 0.66 |
ENSDART00000127843
|
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr10_-_2522588 | 0.66 |
ENSDART00000081926
|
CU856539.1
|
|
| chr3_-_13509283 | 0.66 |
ENSDART00000166832
|
slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr20_-_894374 | 0.66 |
ENSDART00000104725
ENSDART00000142361 |
snx14
|
sorting nexin 14 |
| chr2_+_10878406 | 0.65 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr8_+_48965767 | 0.65 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr22_+_1170294 | 0.65 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr12_+_25223843 | 0.65 |
ENSDART00000077180
ENSDART00000127454 ENSDART00000122665 |
mta3
|
metastasis associated 1 family, member 3 |
| chr12_-_34258384 | 0.65 |
ENSDART00000109196
|
pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr11_+_31324335 | 0.64 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr23_+_4777612 | 0.64 |
ENSDART00000135933
|
vgll4a
|
vestigial-like family member 4a |
| chr17_+_24222190 | 0.64 |
ENSDART00000181698
ENSDART00000189411 |
ehbp1
|
EH domain binding protein 1 |
| chr25_-_34973211 | 0.64 |
ENSDART00000045177
|
cdk10
|
cyclin-dependent kinase 10 |
| chr16_-_36979592 | 0.63 |
ENSDART00000168443
|
snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr23_+_38976201 | 0.62 |
ENSDART00000188090
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr14_-_47391084 | 0.62 |
ENSDART00000159608
|
fstl5
|
follistatin-like 5 |
| chr13_-_41546779 | 0.62 |
ENSDART00000163331
|
pcdh15a
|
protocadherin-related 15a |
| chr20_-_8024582 | 0.62 |
ENSDART00000083898
|
plpp3
|
phospholipid phosphatase 3 |
| chr17_-_33412868 | 0.62 |
ENSDART00000187521
|
BX323819.1
|
|
| chr20_+_18943406 | 0.60 |
ENSDART00000193590
|
mtmr9
|
myotubularin related protein 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of arid3a+arid3b+arid3c
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.6 | 1.9 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.6 | 2.4 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.5 | 4.3 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.5 | 1.4 | GO:0045601 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.5 | 2.8 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.4 | 1.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.4 | 2.1 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.4 | 3.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.4 | 7.2 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.4 | 1.5 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.4 | 1.9 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.4 | 1.5 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.3 | 1.0 | GO:0051230 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.3 | 1.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.3 | 2.0 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.3 | 1.7 | GO:0090133 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.3 | 0.8 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 0.7 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 2.6 | GO:0045453 | bone resorption(GO:0045453) |
| 0.2 | 0.9 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 1.4 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.2 | 3.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.2 | 0.9 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.2 | 0.7 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.2 | 2.4 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 0.6 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 1.1 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.2 | 0.8 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.2 | 1.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 1.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.6 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 1.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.2 | 1.0 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.2 | 1.9 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.2 | 2.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 0.9 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.2 | 2.5 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.2 | 1.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.6 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 0.4 | GO:0015808 | L-alanine transport(GO:0015808) proline transport(GO:0015824) |
| 0.1 | 0.8 | GO:0048714 | positive regulation of gliogenesis(GO:0014015) positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 1.5 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 0.5 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.6 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 1.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 1.1 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 0.4 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 1.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.3 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.1 | 0.5 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 1.1 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 0.3 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.7 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 1.0 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.6 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.3 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 1.0 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 6.9 | GO:0030203 | glycosaminoglycan metabolic process(GO:0030203) |
| 0.1 | 1.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 1.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 1.6 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.3 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.1 | 0.5 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.4 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.1 | 0.7 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.6 | GO:0045719 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.7 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.4 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 0.7 | GO:0031579 | membrane raft organization(GO:0031579) |
| 0.1 | 1.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.5 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 0.4 | GO:1904396 | regulation of neuromuscular junction development(GO:1904396) |
| 0.1 | 5.1 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.1 | 0.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.2 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.1 | 0.8 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.8 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.3 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.4 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.5 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.3 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.0 | 2.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.7 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 1.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 1.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 1.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 1.2 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.6 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 1.1 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.7 | GO:0048885 | neuromast deposition(GO:0048885) |
| 0.0 | 0.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.4 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.8 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.7 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.0 | 1.1 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0051043 | regulation of membrane protein ectodomain proteolysis(GO:0051043) negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.5 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 1.8 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 1.5 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 1.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.7 | GO:0061138 | morphogenesis of a branching epithelium(GO:0061138) |
| 0.0 | 0.7 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 1.1 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 2.1 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.2 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.2 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.8 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.8 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.3 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 1.6 | GO:1903706 | regulation of hemopoiesis(GO:1903706) |
| 0.0 | 0.7 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.0 | 0.3 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.2 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.6 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.3 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 2.5 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.5 | 3.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.4 | 1.3 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.4 | 2.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.4 | 2.2 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.3 | 1.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 0.9 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 1.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 2.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 0.7 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 1.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 2.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.8 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 1.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.7 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.7 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 3.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.6 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 1.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 3.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.5 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 1.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 3.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 1.1 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 3.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 3.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 10.3 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 2.1 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 2.1 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 2.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.7 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.6 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.0 | GO:0031965 | nuclear membrane(GO:0031965) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.5 | 7.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 2.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.3 | 1.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 1.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 3.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 1.5 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.2 | 1.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 0.5 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.2 | 2.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 1.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 0.5 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 1.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.5 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.1 | 0.4 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.7 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 1.5 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 0.5 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 2.5 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 1.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.6 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 0.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 1.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.4 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 1.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.3 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.1 | 1.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.0 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 0.6 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 1.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 2.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.5 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.2 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 6.5 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 3.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.9 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.0 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 1.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.9 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 1.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.6 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 4.2 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 4.9 | GO:0017171 | serine-type peptidase activity(GO:0008236) serine hydrolase activity(GO:0017171) |
| 0.0 | 1.5 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 2.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 3.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 4.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 4.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 2.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 2.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 6.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 3.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.7 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 1.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 3.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 1.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 1.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 1.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |