Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
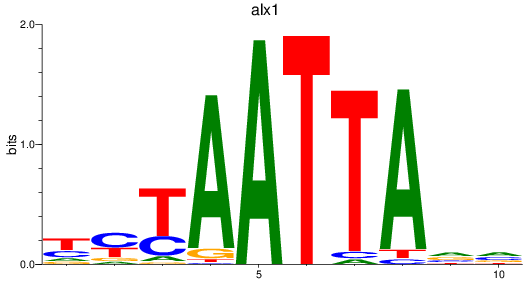
Results for alx1
Z-value: 0.37
Transcription factors associated with alx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
alx1
|
ENSDARG00000062824 | ALX homeobox 1 |
|
alx1
|
ENSDARG00000110530 | ALX homeobox 1 |
|
alx1
|
ENSDARG00000115230 | ALX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| alx1 | dr11_v1_chr18_+_16246806_16246806 | -0.42 | 8.0e-02 | Click! |
Activity profile of alx1 motif
Sorted Z-values of alx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_29509133 | 0.85 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr21_+_25777425 | 0.74 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr8_+_45334255 | 0.72 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr1_-_43905252 | 0.71 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr10_-_21362071 | 0.68 |
ENSDART00000125167
|
avd
|
avidin |
| chr15_-_2519640 | 0.65 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr5_+_37903790 | 0.64 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr10_-_21362320 | 0.64 |
ENSDART00000189789
|
avd
|
avidin |
| chr10_-_25217347 | 0.63 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr22_-_17653143 | 0.62 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr12_-_14143344 | 0.62 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr19_+_15440841 | 0.59 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr16_+_39159752 | 0.58 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr7_-_51773166 | 0.58 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr2_+_41526904 | 0.57 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr14_+_8940326 | 0.56 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr21_-_36453594 | 0.55 |
ENSDART00000193176
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr11_-_1550709 | 0.55 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr5_+_57924611 | 0.53 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr22_-_17652914 | 0.51 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr11_-_35171162 | 0.50 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr5_+_29794058 | 0.49 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr8_+_23355484 | 0.49 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr8_+_11425048 | 0.48 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr19_+_15441022 | 0.46 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr3_+_32416948 | 0.45 |
ENSDART00000157324
ENSDART00000154267 ENSDART00000186094 ENSDART00000155860 ENSDART00000156986 |
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr24_-_2450597 | 0.44 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr5_-_13206878 | 0.43 |
ENSDART00000051666
|
ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr16_-_17347727 | 0.43 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr10_+_16911951 | 0.41 |
ENSDART00000164933
|
UNC13B (1 of many)
|
unc-13 homolog B |
| chr16_-_29387215 | 0.40 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr3_-_30488063 | 0.40 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr8_-_25034411 | 0.39 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr10_-_42147318 | 0.39 |
ENSDART00000134890
|
dusp11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr16_+_13883872 | 0.39 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr17_+_25856671 | 0.39 |
ENSDART00000064817
|
wapla
|
WAPL cohesin release factor a |
| chr5_-_12587053 | 0.38 |
ENSDART00000162780
|
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr14_-_6286966 | 0.38 |
ENSDART00000168174
|
elp1
|
elongator complex protein 1 |
| chr3_-_23575007 | 0.38 |
ENSDART00000155282
ENSDART00000087726 |
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr19_-_82504 | 0.37 |
ENSDART00000027864
ENSDART00000160560 |
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr2_-_49031303 | 0.37 |
ENSDART00000143471
|
cdc34b
|
cell division cycle 34 homolog (S. cerevisiae) b |
| chr25_-_2723682 | 0.37 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr5_+_25733774 | 0.37 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr14_-_33859149 | 0.36 |
ENSDART00000163877
|
ocrl
|
oculocerebrorenal syndrome of Lowe |
| chr7_+_22313533 | 0.36 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr17_-_41798856 | 0.35 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr11_+_12811906 | 0.35 |
ENSDART00000123445
|
rtel1
|
regulator of telomere elongation helicase 1 |
| chr14_-_33945692 | 0.34 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr5_+_44805269 | 0.34 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr9_-_12401871 | 0.34 |
ENSDART00000191901
|
nup35
|
nucleoporin 35 |
| chr8_-_20230802 | 0.34 |
ENSDART00000063400
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr14_-_33481428 | 0.34 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr12_+_22407852 | 0.34 |
ENSDART00000178840
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr2_-_23390779 | 0.33 |
ENSDART00000020136
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr21_-_36453417 | 0.33 |
ENSDART00000018350
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr6_-_41028698 | 0.33 |
ENSDART00000134293
|
trub2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr8_-_20230559 | 0.33 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr17_-_24575893 | 0.32 |
ENSDART00000141914
|
aftphb
|
aftiphilin b |
| chr15_-_43284021 | 0.32 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr16_+_23431189 | 0.32 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr7_+_26649319 | 0.32 |
ENSDART00000173823
ENSDART00000101053 |
tp53i11a
|
tumor protein p53 inducible protein 11a |
| chr20_+_29209926 | 0.31 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr8_+_23356264 | 0.31 |
ENSDART00000145062
|
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr17_-_40956035 | 0.31 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr3_+_41558682 | 0.31 |
ENSDART00000157023
|
card11
|
caspase recruitment domain family, member 11 |
| chr15_-_44052927 | 0.31 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr14_-_26392146 | 0.31 |
ENSDART00000037999
|
b4galt7
|
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) |
| chr14_-_26392475 | 0.30 |
ENSDART00000170614
|
b4galt7
|
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) |
| chr17_+_26722904 | 0.30 |
ENSDART00000114927
|
nrde2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr8_+_3431671 | 0.30 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr1_+_513986 | 0.29 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr4_-_70883663 | 0.29 |
ENSDART00000170959
|
si:ch211-108c17.2
|
si:ch211-108c17.2 |
| chr17_+_16046314 | 0.29 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr24_+_12835935 | 0.29 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr8_-_22538588 | 0.29 |
ENSDART00000144041
|
csde1
|
cold shock domain containing E1, RNA-binding |
| chr14_-_25935167 | 0.29 |
ENSDART00000139855
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr2_-_23391266 | 0.29 |
ENSDART00000159048
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr6_-_50203682 | 0.28 |
ENSDART00000083999
ENSDART00000143050 |
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr2_-_38206034 | 0.28 |
ENSDART00000144518
ENSDART00000137395 |
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr2_-_37312927 | 0.28 |
ENSDART00000141214
|
skila
|
SKI-like proto-oncogene a |
| chr14_+_34492288 | 0.28 |
ENSDART00000144301
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr23_+_32028574 | 0.28 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr15_-_34408777 | 0.28 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr17_+_16090436 | 0.28 |
ENSDART00000136059
ENSDART00000138734 |
znf395a
|
zinc finger protein 395a |
| chr14_+_44860335 | 0.27 |
ENSDART00000091620
ENSDART00000173043 ENSDART00000091625 |
atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr17_+_16046132 | 0.27 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr15_-_33172246 | 0.27 |
ENSDART00000158666
|
nbeab
|
neurobeachin b |
| chr16_+_40340523 | 0.27 |
ENSDART00000102571
|
mettl6
|
methyltransferase like 6 |
| chr20_-_3319642 | 0.27 |
ENSDART00000186743
ENSDART00000123096 |
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr16_+_40340222 | 0.27 |
ENSDART00000190631
|
mettl6
|
methyltransferase like 6 |
| chr11_+_18873619 | 0.27 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr5_+_35786141 | 0.27 |
ENSDART00000022043
ENSDART00000127383 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr1_-_55248496 | 0.27 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr20_+_29209767 | 0.26 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr16_+_54209504 | 0.26 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr14_-_8940499 | 0.26 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr21_+_38638979 | 0.26 |
ENSDART00000143373
|
rbmx2
|
RNA binding motif protein, X-linked 2 |
| chr8_-_22542467 | 0.26 |
ENSDART00000182588
ENSDART00000134542 |
csde1
|
cold shock domain containing E1, RNA-binding |
| chr10_+_17235370 | 0.26 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr6_-_40778294 | 0.25 |
ENSDART00000019845
|
arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr17_-_11495466 | 0.25 |
ENSDART00000020598
|
hnrnpua
|
heterogeneous nuclear ribonucleoprotein Ua |
| chr20_+_29209615 | 0.25 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr2_+_42871831 | 0.25 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr6_-_40922971 | 0.25 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr24_+_19415124 | 0.25 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr14_-_26498196 | 0.25 |
ENSDART00000054175
ENSDART00000145625 ENSDART00000183347 ENSDART00000191084 ENSDART00000191143 |
smad5
|
SMAD family member 5 |
| chr14_+_34490445 | 0.25 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr15_+_1534644 | 0.25 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr13_-_32726178 | 0.25 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr11_+_44804685 | 0.24 |
ENSDART00000163660
|
strn
|
striatin, calmodulin binding protein |
| chr18_+_38807239 | 0.24 |
ENSDART00000184332
|
fam214a
|
family with sequence similarity 214, member A |
| chr21_-_32060993 | 0.24 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr25_+_3759553 | 0.24 |
ENSDART00000180601
ENSDART00000055845 ENSDART00000157050 ENSDART00000153905 |
thoc5
|
THO complex 5 |
| chr18_-_15551360 | 0.24 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr5_-_61797220 | 0.24 |
ENSDART00000079855
|
im:7138535
|
im:7138535 |
| chr21_+_42717424 | 0.24 |
ENSDART00000166936
ENSDART00000172135 |
sh3pxd2b
|
SH3 and PX domains 2B |
| chr7_+_24023653 | 0.24 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr9_+_45428041 | 0.24 |
ENSDART00000193087
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr24_-_5713799 | 0.24 |
ENSDART00000137293
|
dia1b
|
deleted in autism 1b |
| chr17_+_23462972 | 0.24 |
ENSDART00000112959
ENSDART00000192168 |
ankrd1a
|
ankyrin repeat domain 1a (cardiac muscle) |
| chr15_+_21262917 | 0.24 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr7_+_59677273 | 0.23 |
ENSDART00000039535
ENSDART00000132044 |
trmt44
|
tRNA methyltransferase 44 homolog |
| chr3_+_26244353 | 0.23 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr14_+_35428152 | 0.23 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr22_+_34784075 | 0.23 |
ENSDART00000167538
|
lcor
|
ligand dependent nuclear receptor corepressor |
| chr10_-_33297864 | 0.23 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr11_+_31864921 | 0.23 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chrM_+_3803 | 0.22 |
ENSDART00000093596
|
mt-nd1
|
NADH dehydrogenase 1, mitochondrial |
| chr7_-_25133783 | 0.22 |
ENSDART00000173781
ENSDART00000121943 ENSDART00000077219 |
badb
|
BCL2 associated agonist of cell death b |
| chr21_+_31253048 | 0.22 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr21_+_34088377 | 0.22 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr4_-_4261673 | 0.22 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr23_+_31942040 | 0.22 |
ENSDART00000088607
|
nemp1
|
nuclear envelope integral membrane protein 1 |
| chr23_+_9230431 | 0.22 |
ENSDART00000187676
ENSDART00000125122 ENSDART00000190980 ENSDART00000141235 |
acss2
|
acyl-CoA synthetase short chain family member 2 |
| chr7_+_1467863 | 0.22 |
ENSDART00000173433
|
emc4
|
ER membrane protein complex subunit 4 |
| chr11_+_2649891 | 0.22 |
ENSDART00000093052
|
si:ch211-160o17.4
|
si:ch211-160o17.4 |
| chr3_+_19248973 | 0.22 |
ENSDART00000174668
|
smarca4a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4a |
| chr24_-_25166720 | 0.22 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr19_+_20793237 | 0.22 |
ENSDART00000014774
|
txnl4a
|
thioredoxin-like 4A |
| chr22_-_14161309 | 0.21 |
ENSDART00000133365
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr12_+_33361948 | 0.21 |
ENSDART00000124982
|
fasn
|
fatty acid synthase |
| chr6_+_29693492 | 0.21 |
ENSDART00000114172
|
pde6d
|
phosphodiesterase 6D, cGMP-specific, rod, delta |
| chr20_+_32523576 | 0.21 |
ENSDART00000147319
|
scml4
|
Scm polycomb group protein like 4 |
| chr23_+_20563779 | 0.21 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr10_-_35103208 | 0.21 |
ENSDART00000192734
|
zgc:110006
|
zgc:110006 |
| chr2_-_26596794 | 0.21 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr19_+_6990970 | 0.21 |
ENSDART00000158758
ENSDART00000160482 ENSDART00000193566 |
kifc1
|
kinesin family member C1 |
| chr10_+_11261576 | 0.21 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr10_+_7718156 | 0.21 |
ENSDART00000189101
|
ggcx
|
gamma-glutamyl carboxylase |
| chr23_+_33963619 | 0.21 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr3_+_18579806 | 0.20 |
ENSDART00000180967
ENSDART00000089765 |
arhgap17b
|
Rho GTPase activating protein 17b |
| chr8_+_3820134 | 0.20 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr12_-_4408828 | 0.20 |
ENSDART00000152447
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr19_+_9032073 | 0.20 |
ENSDART00000127755
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr20_+_16639848 | 0.20 |
ENSDART00000063944
ENSDART00000152359 |
tmem30ab
|
transmembrane protein 30Ab |
| chr5_-_68093169 | 0.20 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr19_+_7549854 | 0.20 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr16_-_28658341 | 0.20 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr8_+_28452738 | 0.20 |
ENSDART00000062706
|
tmem189
|
transmembrane protein 189 |
| chr13_-_35808904 | 0.20 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr9_-_27398369 | 0.20 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr7_-_59165640 | 0.20 |
ENSDART00000170853
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr24_+_1023839 | 0.20 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr24_+_36204028 | 0.20 |
ENSDART00000063832
ENSDART00000155260 |
rbbp8
|
retinoblastoma binding protein 8 |
| chr9_+_34397516 | 0.20 |
ENSDART00000011304
ENSDART00000192973 |
med14
|
mediator complex subunit 14 |
| chr24_-_24724233 | 0.20 |
ENSDART00000127044
ENSDART00000012399 |
armc1
|
armadillo repeat containing 1 |
| chr22_-_20924564 | 0.19 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr2_+_1714640 | 0.19 |
ENSDART00000086761
ENSDART00000111613 |
adgrl2b.1
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr17_-_25303486 | 0.19 |
ENSDART00000162235
|
ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr3_+_18807006 | 0.19 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr21_+_34088110 | 0.19 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr8_+_50953776 | 0.19 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr19_+_26922780 | 0.19 |
ENSDART00000187396
ENSDART00000188978 |
nelfe
|
negative elongation factor complex member E |
| chr6_-_12172424 | 0.19 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr21_-_13662237 | 0.19 |
ENSDART00000091647
ENSDART00000151547 |
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr2_-_37098785 | 0.19 |
ENSDART00000003670
|
zgc:101744
|
zgc:101744 |
| chr6_+_36839509 | 0.19 |
ENSDART00000190605
ENSDART00000104160 |
zgc:110788
|
zgc:110788 |
| chr15_-_37543591 | 0.18 |
ENSDART00000180400
|
kmt2bb
|
lysine (K)-specific methyltransferase 2Bb |
| chr9_+_35014728 | 0.18 |
ENSDART00000100700
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr7_-_54217547 | 0.18 |
ENSDART00000162777
ENSDART00000188268 ENSDART00000165875 |
csnk1g1
|
casein kinase 1, gamma 1 |
| chr3_+_45364849 | 0.18 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr3_+_27786601 | 0.18 |
ENSDART00000086994
|
nat15
|
N-acetyltransferase 15 (GCN5-related, putative) |
| chr13_-_28308138 | 0.18 |
ENSDART00000045351
|
lbx1a
|
ladybird homeobox 1a |
| chr16_+_33163858 | 0.18 |
ENSDART00000101943
|
rragca
|
Ras-related GTP binding Ca |
| chr7_+_19483277 | 0.18 |
ENSDART00000173750
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr9_-_14683574 | 0.18 |
ENSDART00000144022
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr11_-_16115804 | 0.17 |
ENSDART00000143436
ENSDART00000157928 |
rpf1
|
ribosome production factor 1 homolog |
| chr16_-_25368048 | 0.17 |
ENSDART00000132445
|
rbfa
|
ribosome binding factor A |
| chr19_+_46158078 | 0.17 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr4_+_13931578 | 0.17 |
ENSDART00000142466
|
pphln1
|
periphilin 1 |
| chr10_+_42521943 | 0.17 |
ENSDART00000010420
ENSDART00000075303 |
actr1
|
ARP1 actin related protein 1, centractin |
| chr4_-_16124417 | 0.17 |
ENSDART00000128079
ENSDART00000077664 |
atp2b1a
|
ATPase plasma membrane Ca2+ transporting 1a |
| chr8_-_23776399 | 0.17 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr9_+_8396755 | 0.17 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr10_-_11323134 | 0.17 |
ENSDART00000039145
|
inip
|
ints3 and nabp interacting protein |
| chr12_+_8822717 | 0.17 |
ENSDART00000021628
|
reep3b
|
receptor accessory protein 3b |
| chr10_+_7593185 | 0.17 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr17_+_22587356 | 0.16 |
ENSDART00000157328
|
birc6
|
baculoviral IAP repeat containing 6 |
| chr13_-_23095006 | 0.16 |
ENSDART00000089242
|
kif1bp
|
kif1 binding protein |
| chr16_+_19637384 | 0.16 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of alx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.6 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.6 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.1 | 0.6 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 0.4 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.4 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.1 | 0.6 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.4 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 0.3 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 1.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.6 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
| 0.1 | 0.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.2 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 0.3 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.3 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 0.5 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.0 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.2 | GO:2000389 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.3 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.2 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.2 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.2 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.2 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.2 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.2 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.7 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.4 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.6 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.2 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.1 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.0 | 0.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.2 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.1 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.2 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.0 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.3 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.1 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.3 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 0.6 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 0.5 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.1 | 0.3 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.3 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.3 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.2 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.9 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.3 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.6 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.4 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 1.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.2 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.2 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.2 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 0.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.8 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.2 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.6 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0072591 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.6 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |