Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
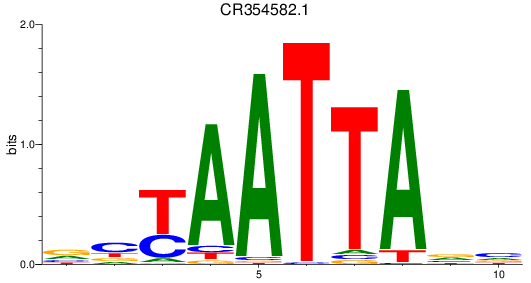
Results for CR354582.1
Z-value: 0.60
Transcription factors associated with CR354582.1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CR354582.1
|
ENSDARG00000110598 | ENSDARG00000110598 |
Activity profile of CR354582.1 motif
Sorted Z-values of CR354582.1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_23426339 | 2.39 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr9_-_35633827 | 2.32 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr2_+_6253246 | 1.80 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr16_-_42056137 | 1.60 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr11_+_18183220 | 1.25 |
ENSDART00000113468
|
LO018315.10
|
|
| chr4_+_11464255 | 1.16 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr24_-_37640705 | 1.09 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr3_+_28860283 | 0.95 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr1_-_18811517 | 0.83 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr24_-_39518599 | 0.82 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr2_+_8779164 | 0.75 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr18_-_40708537 | 0.73 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr22_+_4488454 | 0.71 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr11_+_33818179 | 0.70 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr1_+_55755304 | 0.63 |
ENSDART00000144983
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr9_+_50001746 | 0.58 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr24_+_40860320 | 0.57 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr8_-_23780334 | 0.57 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr17_+_24821627 | 0.56 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr20_-_16171297 | 0.51 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr16_-_43344859 | 0.51 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr20_-_37813863 | 0.47 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr12_-_48477031 | 0.47 |
ENSDART00000105176
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr3_+_18398876 | 0.47 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr6_+_50381347 | 0.47 |
ENSDART00000055504
|
cyc1
|
cytochrome c-1 |
| chr3_+_17537352 | 0.44 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr7_-_30174882 | 0.44 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr17_+_37227936 | 0.43 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr16_+_33902006 | 0.43 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr10_-_21362071 | 0.42 |
ENSDART00000125167
|
avd
|
avidin |
| chr8_+_25034544 | 0.41 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr10_-_21362320 | 0.41 |
ENSDART00000189789
|
avd
|
avidin |
| chr24_+_37640626 | 0.39 |
ENSDART00000008047
|
wdr24
|
WD repeat domain 24 |
| chr2_+_50608099 | 0.39 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr12_+_20352400 | 0.38 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr19_-_5103141 | 0.38 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr5_-_25733745 | 0.38 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr8_-_21142550 | 0.38 |
ENSDART00000143192
ENSDART00000186820 ENSDART00000135938 |
cpt2
|
carnitine palmitoyltransferase 2 |
| chr11_-_41220794 | 0.36 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr25_-_10503043 | 0.35 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr21_-_5205617 | 0.35 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr6_+_50381665 | 0.35 |
ENSDART00000141128
|
cyc1
|
cytochrome c-1 |
| chr23_+_44374041 | 0.34 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr12_+_22580579 | 0.34 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr3_+_32365811 | 0.34 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr2_-_15324837 | 0.32 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr10_-_26512993 | 0.32 |
ENSDART00000188549
ENSDART00000193316 |
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr1_-_55248496 | 0.32 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr10_-_26512742 | 0.31 |
ENSDART00000135951
|
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr19_+_42432625 | 0.31 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr5_+_6954162 | 0.30 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr11_-_15296805 | 0.28 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr19_-_5103313 | 0.27 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr9_-_19161982 | 0.27 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr21_-_32781612 | 0.27 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr5_-_63302944 | 0.26 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr10_-_20696518 | 0.26 |
ENSDART00000064581
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr6_+_49926115 | 0.25 |
ENSDART00000018523
|
ahcy
|
adenosylhomocysteinase |
| chr11_+_33312601 | 0.25 |
ENSDART00000188024
|
cntnap5l
|
contactin associated protein-like 5 like |
| chr22_-_20166660 | 0.25 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr6_-_7720332 | 0.24 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr6_-_55585423 | 0.23 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr10_-_32494304 | 0.23 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr10_-_32494499 | 0.22 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr19_+_31904836 | 0.21 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr8_-_37249813 | 0.21 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr14_+_45471642 | 0.21 |
ENSDART00000126979
ENSDART00000172952 ENSDART00000173284 |
ubxn1
|
UBX domain protein 1 |
| chr5_-_9625459 | 0.20 |
ENSDART00000143347
|
sh2b3
|
SH2B adaptor protein 3 |
| chr17_+_16046132 | 0.20 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr3_-_31079186 | 0.20 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr10_+_39212898 | 0.18 |
ENSDART00000159501
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr17_+_16046314 | 0.18 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr11_-_6048490 | 0.18 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr25_-_13490744 | 0.18 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr16_+_42471455 | 0.18 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr15_-_47848544 | 0.18 |
ENSDART00000098711
|
eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr20_-_9436521 | 0.17 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr15_-_16177603 | 0.16 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr8_-_37249991 | 0.16 |
ENSDART00000189275
ENSDART00000178556 |
rbm39b
|
RNA binding motif protein 39b |
| chr10_+_39212601 | 0.15 |
ENSDART00000168778
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr22_-_10586191 | 0.15 |
ENSDART00000148418
|
si:dkey-42i9.16
|
si:dkey-42i9.16 |
| chr9_-_22057658 | 0.15 |
ENSDART00000101944
|
crygmxl1
|
crystallin, gamma MX, like 1 |
| chr9_-_50000144 | 0.15 |
ENSDART00000123416
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr8_+_45334255 | 0.15 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr20_-_45060241 | 0.14 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr14_-_2933185 | 0.14 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr17_+_24722646 | 0.13 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr1_-_29139141 | 0.13 |
ENSDART00000075546
ENSDART00000133246 |
hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr1_+_21731382 | 0.12 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr19_-_26736336 | 0.12 |
ENSDART00000109258
ENSDART00000182802 |
csnk2b
|
casein kinase 2, beta polypeptide |
| chr9_-_3934963 | 0.12 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr24_+_16547035 | 0.11 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr20_-_9462433 | 0.10 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr25_+_14507567 | 0.10 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr16_+_5774977 | 0.10 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr10_+_17714866 | 0.10 |
ENSDART00000039969
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr19_+_390298 | 0.10 |
ENSDART00000136361
|
sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr13_+_38817871 | 0.10 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr1_+_52929185 | 0.10 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr19_+_43297546 | 0.09 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr3_-_12930217 | 0.09 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr20_-_9095105 | 0.09 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr16_+_17389116 | 0.09 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr7_-_23768234 | 0.08 |
ENSDART00000173981
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr22_+_20427170 | 0.08 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr21_-_39177564 | 0.08 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr22_-_13350240 | 0.08 |
ENSDART00000154095
ENSDART00000155118 |
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr3_+_34919810 | 0.08 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr19_+_31585917 | 0.07 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr22_-_10121880 | 0.07 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr6_+_52873822 | 0.07 |
ENSDART00000103138
|
or137-3
|
odorant receptor, family H, subfamily 137, member 3 |
| chr9_-_20372977 | 0.07 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr14_+_25817628 | 0.07 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr24_-_35282568 | 0.07 |
ENSDART00000167406
ENSDART00000088609 |
sntg1
|
syntrophin, gamma 1 |
| chr5_+_66433287 | 0.07 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr13_+_38814521 | 0.07 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr14_+_45406299 | 0.06 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr18_+_20560442 | 0.06 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr24_+_38306010 | 0.06 |
ENSDART00000143184
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr13_+_27232848 | 0.06 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr12_-_35830625 | 0.06 |
ENSDART00000180028
|
CU459056.1
|
|
| chr3_+_17933553 | 0.05 |
ENSDART00000167731
ENSDART00000165644 |
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr19_+_9174166 | 0.05 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr17_-_40956035 | 0.05 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr15_+_31344472 | 0.05 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr23_+_26946744 | 0.05 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr23_-_1658980 | 0.05 |
ENSDART00000186227
|
CU693481.1
|
|
| chr3_-_50443607 | 0.05 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr24_+_12894282 | 0.05 |
ENSDART00000061301
|
si:dkeyp-28d2.4
|
si:dkeyp-28d2.4 |
| chr20_+_535458 | 0.04 |
ENSDART00000152453
|
dse
|
dermatan sulfate epimerase |
| chr5_-_12219572 | 0.04 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr24_+_6107901 | 0.04 |
ENSDART00000156419
|
si:ch211-37e10.2
|
si:ch211-37e10.2 |
| chr5_-_27438812 | 0.04 |
ENSDART00000078755
|
drd7
|
dopamine receptor D7 |
| chr17_+_21546993 | 0.04 |
ENSDART00000182387
|
chst15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr14_+_34951202 | 0.04 |
ENSDART00000047524
ENSDART00000115105 |
il12ba
|
interleukin 12Ba |
| chr8_-_24252933 | 0.04 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr14_+_40874608 | 0.04 |
ENSDART00000168448
|
si:ch211-106m9.1
|
si:ch211-106m9.1 |
| chr3_-_46811611 | 0.04 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr25_-_25384045 | 0.04 |
ENSDART00000150631
|
zgc:123278
|
zgc:123278 |
| chr8_-_12867128 | 0.04 |
ENSDART00000142201
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr1_-_5455498 | 0.04 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr21_+_32820175 | 0.04 |
ENSDART00000076903
|
adra2db
|
adrenergic, alpha-2D-, receptor b |
| chr11_+_24703108 | 0.04 |
ENSDART00000159173
|
gpr25
|
G protein-coupled receptor 25 |
| chr23_+_44741500 | 0.03 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr12_-_33357655 | 0.03 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr17_+_23298928 | 0.03 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr9_-_50001606 | 0.03 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr3_-_46817499 | 0.03 |
ENSDART00000013717
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr4_+_12612145 | 0.03 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr11_-_45138857 | 0.03 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr6_+_9289802 | 0.03 |
ENSDART00000188650
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr23_-_6660985 | 0.03 |
ENSDART00000162405
|
CR450824.2
|
|
| chr6_-_6487876 | 0.03 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr6_-_40352215 | 0.03 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr9_+_22364997 | 0.03 |
ENSDART00000188054
ENSDART00000046116 |
crygs3
|
crystallin, gamma S3 |
| chr19_-_20113696 | 0.03 |
ENSDART00000188813
|
npy
|
neuropeptide Y |
| chr8_-_12867434 | 0.03 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr19_-_32710922 | 0.03 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr21_+_11244068 | 0.03 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr18_+_50461981 | 0.03 |
ENSDART00000158761
|
CU896640.1
|
|
| chr10_-_34002185 | 0.03 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr19_-_657439 | 0.03 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr22_-_27709692 | 0.03 |
ENSDART00000172458
|
CR547131.1
|
|
| chr4_-_4387012 | 0.02 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr2_-_30668580 | 0.02 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr22_-_12746539 | 0.02 |
ENSDART00000175374
|
plcd4a
|
phospholipase C, delta 4a |
| chr23_+_28582865 | 0.02 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr8_+_20776654 | 0.02 |
ENSDART00000135850
|
nfic
|
nuclear factor I/C |
| chr18_+_48423973 | 0.02 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr4_-_71912457 | 0.02 |
ENSDART00000179685
|
si:dkey-92c21.1
|
si:dkey-92c21.1 |
| chr21_-_35419486 | 0.02 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr14_+_33329420 | 0.02 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr23_+_7548797 | 0.02 |
ENSDART00000006765
|
tm9sf4
|
transmembrane 9 superfamily protein member 4 |
| chr11_+_30244356 | 0.02 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr20_-_46362606 | 0.02 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr12_+_10631266 | 0.02 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr22_+_17205608 | 0.02 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr14_+_30795559 | 0.02 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr23_-_19230627 | 0.02 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr11_+_38280454 | 0.02 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr1_-_40911332 | 0.02 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr10_+_25726694 | 0.02 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr19_-_30524952 | 0.02 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr5_+_30624183 | 0.02 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr12_-_19007834 | 0.01 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr7_+_48667081 | 0.01 |
ENSDART00000083473
|
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr15_-_22074315 | 0.01 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr19_+_10339538 | 0.01 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr20_-_40755614 | 0.01 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr2_-_23778180 | 0.01 |
ENSDART00000136782
|
si:dkey-24c2.7
|
si:dkey-24c2.7 |
| chr11_-_3865472 | 0.01 |
ENSDART00000161426
|
gata2a
|
GATA binding protein 2a |
| chr3_-_23643751 | 0.01 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr17_+_8799451 | 0.01 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr18_-_2433011 | 0.01 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr17_-_23616626 | 0.01 |
ENSDART00000104730
|
ifit14
|
interferon-induced protein with tetratricopeptide repeats 14 |
| chr10_+_40660772 | 0.01 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr6_+_40922572 | 0.01 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr10_-_40421050 | 0.01 |
ENSDART00000141834
ENSDART00000164847 |
taar20w
|
trace amine associated receptor 20w |
| chr10_-_11385155 | 0.01 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr19_-_5265155 | 0.00 |
ENSDART00000145003
|
prf1.3
|
perforin 1.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CR354582.1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.7 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.4 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 0.4 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.8 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.9 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 1.0 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.7 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 2.4 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.1 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.0 | 0.8 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex I biogenesis(GO:0097031) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) post-translational protein modification(GO:0043687) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.0 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.3 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.8 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.2 | 3.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 0.8 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 0.7 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.3 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.4 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.2 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.0 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.0 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |