Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for CABZ01093502.2
Z-value: 0.11
Transcription factors associated with CABZ01093502.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CABZ01093502.2
|
ENSDARG00000105053 | developing brain homeobox 2 |
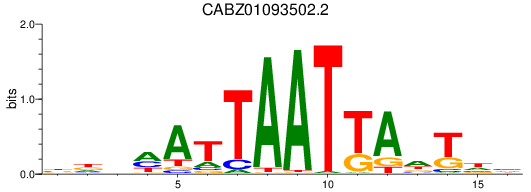
Activity profile of CABZ01093502.2 motif
Sorted Z-values of CABZ01093502.2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_3915770 | 0.34 |
ENSDART00000159322
|
si:ch73-111k22.3
|
si:ch73-111k22.3 |
| chr2_-_9818640 | 0.29 |
ENSDART00000139499
ENSDART00000165548 ENSDART00000012442 ENSDART00000046587 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr3_+_26145013 | 0.27 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr11_-_2838699 | 0.22 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr15_-_46779934 | 0.17 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr6_+_13117598 | 0.15 |
ENSDART00000104744
|
casp8l1
|
caspase 8, apoptosis-related cysteine peptidase, like 1 |
| chr2_+_19522082 | 0.15 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr2_+_14992879 | 0.14 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr23_-_6522099 | 0.11 |
ENSDART00000092214
ENSDART00000183380 ENSDART00000138020 |
bmp7b
|
bone morphogenetic protein 7b |
| chr9_-_9415000 | 0.11 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr20_-_9095105 | 0.11 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr20_+_25586099 | 0.11 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr23_-_16485190 | 0.11 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr10_-_5847655 | 0.10 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr22_-_26865361 | 0.10 |
ENSDART00000182504
|
hmox2a
|
heme oxygenase 2a |
| chr11_+_30057762 | 0.10 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr12_-_10409961 | 0.10 |
ENSDART00000149521
ENSDART00000052001 |
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr1_-_19502322 | 0.10 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr2_+_19578446 | 0.10 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr11_-_1509773 | 0.10 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr22_-_26865181 | 0.09 |
ENSDART00000138311
|
hmox2a
|
heme oxygenase 2a |
| chr4_+_9669717 | 0.09 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr24_-_38657683 | 0.09 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr23_+_22656477 | 0.08 |
ENSDART00000009337
ENSDART00000133322 |
eno1a
|
enolase 1a, (alpha) |
| chr4_-_9891874 | 0.08 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr19_+_1688727 | 0.08 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr20_+_11731039 | 0.08 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr8_+_52637507 | 0.07 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr23_-_37113396 | 0.07 |
ENSDART00000102886
ENSDART00000134461 |
zgc:193690
|
zgc:193690 |
| chr20_+_40457599 | 0.07 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr11_+_40812590 | 0.07 |
ENSDART00000186690
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr15_-_21014270 | 0.05 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr23_-_37113215 | 0.05 |
ENSDART00000146835
|
zgc:193690
|
zgc:193690 |
| chr22_-_36530902 | 0.05 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr17_+_44780166 | 0.05 |
ENSDART00000156260
|
tmem63c
|
transmembrane protein 63C |
| chr12_-_28363111 | 0.05 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr25_+_3318192 | 0.05 |
ENSDART00000146154
|
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr24_-_6078222 | 0.05 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr11_-_29737088 | 0.05 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr2_+_2223837 | 0.05 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr1_-_31171242 | 0.04 |
ENSDART00000190294
|
kcnq5b
|
potassium voltage-gated channel, KQT-like subfamily, member 5b |
| chr20_+_41756996 | 0.04 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr14_-_413273 | 0.04 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr25_-_6011034 | 0.04 |
ENSDART00000075197
ENSDART00000136054 |
snx22
|
sorting nexin 22 |
| chr16_+_23303859 | 0.04 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr1_+_51721851 | 0.03 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr14_-_34771371 | 0.03 |
ENSDART00000160598
ENSDART00000150413 ENSDART00000168910 |
ablim3
|
actin binding LIM protein family, member 3 |
| chr24_-_7995960 | 0.03 |
ENSDART00000186594
|
bloc1s5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr13_-_12602920 | 0.03 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr5_-_42904329 | 0.03 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr8_-_30204650 | 0.02 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr6_-_35046735 | 0.02 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr5_+_34623107 | 0.02 |
ENSDART00000184126
|
enc1
|
ectodermal-neural cortex 1 |
| chr6_-_58764672 | 0.02 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr21_+_28478663 | 0.02 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr22_-_24818066 | 0.02 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr10_-_20339113 | 0.02 |
ENSDART00000144378
ENSDART00000098600 |
zmat4b
|
zinc finger, matrin-type 4b |
| chr4_-_49582758 | 0.02 |
ENSDART00000180834
ENSDART00000187608 |
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr20_-_46128590 | 0.01 |
ENSDART00000123744
|
taar1b
|
trace amine associated receptor 1b |
| chr9_-_51436377 | 0.01 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr19_+_33093577 | 0.01 |
ENSDART00000180317
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr2_-_59327299 | 0.01 |
ENSDART00000133734
|
ftr36
|
finTRIM family, member 36 |
| chr3_-_19368435 | 0.01 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr19_+_43780970 | 0.01 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr8_-_13184989 | 0.01 |
ENSDART00000135738
|
zgc:194990
|
zgc:194990 |
| chr17_-_10122204 | 0.01 |
ENSDART00000160751
|
BX088587.1
|
|
| chr16_-_38118003 | 0.01 |
ENSDART00000058667
|
si:dkey-23o4.6
|
si:dkey-23o4.6 |
| chr1_+_58242498 | 0.00 |
ENSDART00000149091
|
ggt1l2.2
|
gamma-glutamyltransferase 1 like 2.2 |
| chr8_+_6576940 | 0.00 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr18_+_30847237 | 0.00 |
ENSDART00000012374
|
foxf1
|
forkhead box F1 |
| chr5_-_30620625 | 0.00 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr5_+_71802014 | 0.00 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr15_-_17618800 | 0.00 |
ENSDART00000157185
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr2_+_55984788 | 0.00 |
ENSDART00000183599
|
nmrk2
|
nicotinamide riboside kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CABZ01093502.2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.3 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |