Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
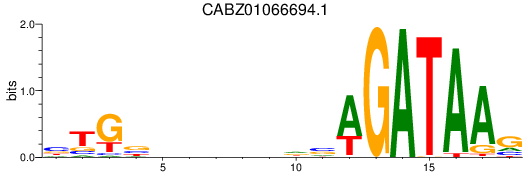
Results for CABZ01066694.1
Z-value: 0.37
Transcription factors associated with CABZ01066694.1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CABZ01066694.1
|
ENSDARG00000110178 | ENSDARG00000110178 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CABZ01066694.1 | dr11_v1_chr6_+_168962_168962 | 0.18 | 4.9e-01 | Click! |
Activity profile of CABZ01066694.1 motif
Sorted Z-values of CABZ01066694.1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_10920577 | 0.85 |
ENSDART00000165630
|
ecm1a
|
extracellular matrix protein 1a |
| chr19_-_27578929 | 0.75 |
ENSDART00000177368
|
si:dkeyp-46h3.3
|
si:dkeyp-46h3.3 |
| chr3_-_3428938 | 0.61 |
ENSDART00000179811
ENSDART00000115282 ENSDART00000192263 ENSDART00000046454 |
A2ML1 (1 of many)
|
zgc:171445 |
| chr19_+_48102560 | 0.58 |
ENSDART00000164464
|
utp18
|
UTP18 small subunit (SSU) processome component |
| chr3_-_3413669 | 0.56 |
ENSDART00000113517
ENSDART00000179861 ENSDART00000115331 |
zgc:171446
|
zgc:171446 |
| chr19_-_27564458 | 0.54 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr2_-_42375275 | 0.53 |
ENSDART00000026339
|
gtpbp4
|
GTP binding protein 4 |
| chr7_+_12931396 | 0.52 |
ENSDART00000184664
|
fth1a
|
ferritin, heavy polypeptide 1a |
| chr21_+_198269 | 0.51 |
ENSDART00000193485
ENSDART00000163696 |
gsdf
|
gonadal somatic cell derived factor |
| chr3_-_1387292 | 0.50 |
ENSDART00000163535
|
ddx47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr4_+_27100531 | 0.43 |
ENSDART00000115200
|
alg12
|
ALG12, alpha-1,6-mannosyltransferase |
| chr5_+_67971627 | 0.41 |
ENSDART00000144879
|
mtif3
|
mitochondrial translational initiation factor 3 |
| chr15_+_34592215 | 0.41 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr16_-_24668620 | 0.37 |
ENSDART00000012807
|
pon3.2
|
paraoxonase 3, tandem duplicate 2 |
| chr23_+_22658700 | 0.29 |
ENSDART00000192248
|
eno1a
|
enolase 1a, (alpha) |
| chr20_+_33981946 | 0.28 |
ENSDART00000131775
|
si:dkey-51e6.1
|
si:dkey-51e6.1 |
| chr16_-_45069882 | 0.27 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr2_+_8779164 | 0.27 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr15_-_24588649 | 0.26 |
ENSDART00000060469
|
gemin4
|
gem (nuclear organelle) associated protein 4 |
| chr2_+_16160906 | 0.24 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr25_+_16098620 | 0.23 |
ENSDART00000142564
ENSDART00000165598 |
far1
|
fatty acyl CoA reductase 1 |
| chr20_-_51727860 | 0.22 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr20_-_26066020 | 0.22 |
ENSDART00000078559
|
myct1a
|
myc target 1a |
| chr17_-_25831569 | 0.21 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr25_+_2415131 | 0.20 |
ENSDART00000162210
|
zmp:0000000932
|
zmp:0000000932 |
| chr24_-_11050727 | 0.20 |
ENSDART00000145466
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr25_+_35062353 | 0.20 |
ENSDART00000089844
|
zgc:113983
|
zgc:113983 |
| chr13_-_5065539 | 0.20 |
ENSDART00000141790
|
abcg2c
|
ATP-binding cassette, sub-family G (WHITE), member 2c |
| chr16_+_45930962 | 0.20 |
ENSDART00000124689
ENSDART00000041811 |
otud7b
|
OTU deubiquitinase 7B |
| chr16_-_42303856 | 0.20 |
ENSDART00000180030
|
ppox
|
protoporphyrinogen oxidase |
| chr9_-_7652792 | 0.19 |
ENSDART00000137957
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr15_-_39969988 | 0.19 |
ENSDART00000146054
|
rps5
|
ribosomal protein S5 |
| chr10_+_15255198 | 0.18 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr21_-_25756119 | 0.18 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr24_+_42129775 | 0.17 |
ENSDART00000181576
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr10_+_15255012 | 0.17 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr24_+_18286427 | 0.17 |
ENSDART00000055443
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr3_+_59411956 | 0.17 |
ENSDART00000166982
|
sec14l1
|
SEC14-like lipid binding 1 |
| chr1_+_24557414 | 0.16 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr1_-_23274038 | 0.16 |
ENSDART00000181658
|
rfc1
|
replication factor C (activator 1) 1 |
| chr14_-_10371638 | 0.15 |
ENSDART00000003879
|
fam199x
|
family with sequence similarity 199, X-linked |
| chr10_-_17484762 | 0.15 |
ENSDART00000137905
ENSDART00000007961 |
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr1_+_25585 | 0.15 |
ENSDART00000161139
|
CU651657.1
|
|
| chr21_-_11834452 | 0.14 |
ENSDART00000081652
|
rfesd
|
Rieske (Fe-S) domain containing |
| chr24_-_21498802 | 0.14 |
ENSDART00000181235
ENSDART00000153695 |
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr21_+_39432248 | 0.14 |
ENSDART00000179938
|
pafah1b1b
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b |
| chr23_+_4689626 | 0.14 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr6_-_49078263 | 0.13 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr21_-_12030654 | 0.13 |
ENSDART00000139145
|
tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr10_+_39476432 | 0.13 |
ENSDART00000190375
ENSDART00000183954 |
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr3_+_32577994 | 0.12 |
ENSDART00000180643
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr1_+_9082417 | 0.12 |
ENSDART00000146303
|
cacng3a
|
calcium channel, voltage-dependent, gamma subunit 3a |
| chr8_-_7637626 | 0.12 |
ENSDART00000123882
ENSDART00000040672 |
mecp2
|
methyl CpG binding protein 2 |
| chr15_-_21837207 | 0.12 |
ENSDART00000089953
|
sik2b
|
salt-inducible kinase 2b |
| chr8_-_37249813 | 0.12 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr8_-_46045172 | 0.12 |
ENSDART00000135138
|
si:ch211-119d14.3
|
si:ch211-119d14.3 |
| chr22_-_30935510 | 0.11 |
ENSDART00000133335
|
si:dkey-49n23.1
|
si:dkey-49n23.1 |
| chr11_+_41981959 | 0.10 |
ENSDART00000055707
|
her15.1
|
hairy and enhancer of split-related 15, tandem duplicate 1 |
| chr8_-_37249991 | 0.10 |
ENSDART00000189275
ENSDART00000178556 |
rbm39b
|
RNA binding motif protein 39b |
| chr10_-_28368695 | 0.10 |
ENSDART00000179416
|
cltca
|
clathrin, heavy chain a (Hc) |
| chr16_+_29630965 | 0.10 |
ENSDART00000185820
ENSDART00000192105 ENSDART00000153683 ENSDART00000186713 |
VPS72
|
si:ch211-203d17.1 |
| chr14_+_17197132 | 0.10 |
ENSDART00000054598
|
rtn4rl2b
|
reticulon 4 receptor-like 2b |
| chr7_+_8324506 | 0.10 |
ENSDART00000168110
|
si:dkey-185m8.2
|
si:dkey-185m8.2 |
| chr18_+_14633974 | 0.10 |
ENSDART00000133834
|
vps9d1
|
VPS9 domain containing 1 |
| chr24_-_39858710 | 0.10 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr15_+_9294620 | 0.09 |
ENSDART00000133588
ENSDART00000140009 |
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr3_-_5555500 | 0.09 |
ENSDART00000176133
ENSDART00000057464 |
trim35-31
|
tripartite motif containing 35-31 |
| chr10_-_32558917 | 0.09 |
ENSDART00000128888
ENSDART00000143301 |
mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr7_+_5052336 | 0.09 |
ENSDART00000133393
|
si:ch211-272h9.6
|
si:ch211-272h9.6 |
| chr12_+_7497882 | 0.08 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr4_-_1839352 | 0.08 |
ENSDART00000189215
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr15_+_36115955 | 0.08 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr18_+_34478959 | 0.08 |
ENSDART00000059394
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr10_+_36345176 | 0.08 |
ENSDART00000099397
|
or105-1
|
odorant receptor, family C, subfamily 105, member 1 |
| chr20_+_6590220 | 0.08 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr7_-_38638809 | 0.07 |
ENSDART00000144341
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr9_+_52613820 | 0.07 |
ENSDART00000168753
ENSDART00000165580 ENSDART00000164769 |
si:ch211-241j8.2
|
si:ch211-241j8.2 |
| chr16_-_30931651 | 0.07 |
ENSDART00000132643
|
slc45a4
|
solute carrier family 45, member 4 |
| chr7_+_41748693 | 0.07 |
ENSDART00000174379
ENSDART00000052168 |
hrh3
|
histamine receptor H3 |
| chr4_-_1839192 | 0.07 |
ENSDART00000003790
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr5_-_69523816 | 0.07 |
ENSDART00000112692
|
si:ch211-157p22.10
|
si:ch211-157p22.10 |
| chr3_-_20091964 | 0.06 |
ENSDART00000029386
ENSDART00000020253 ENSDART00000124326 |
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr19_+_37509638 | 0.06 |
ENSDART00000139999
|
si:ch211-250g4.3
|
si:ch211-250g4.3 |
| chr8_+_21353878 | 0.06 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr3_+_52737565 | 0.06 |
ENSDART00000108639
|
gmip
|
GEM interacting protein |
| chr5_-_30088070 | 0.06 |
ENSDART00000078115
ENSDART00000134209 |
sdhda
|
succinate dehydrogenase complex, subunit D, integral membrane protein a |
| chr5_+_42388464 | 0.06 |
ENSDART00000191596
|
BX548073.13
|
|
| chr10_+_41199660 | 0.06 |
ENSDART00000125314
|
adrb3b
|
adrenoceptor beta 3b |
| chr16_-_30931468 | 0.06 |
ENSDART00000187838
|
slc45a4
|
solute carrier family 45, member 4 |
| chr21_+_25935845 | 0.06 |
ENSDART00000141149
|
caln2
|
calneuron 2 |
| chr2_-_32768951 | 0.06 |
ENSDART00000004712
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr15_+_16886196 | 0.06 |
ENSDART00000139296
ENSDART00000049196 |
gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr3_+_17346502 | 0.06 |
ENSDART00000187786
ENSDART00000131584 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr4_+_39742119 | 0.05 |
ENSDART00000176004
|
CR749167.2
|
|
| chr2_+_21486529 | 0.05 |
ENSDART00000047468
|
inhbab
|
inhibin, beta Ab |
| chr3_-_11203689 | 0.05 |
ENSDART00000166328
|
BX323882.1
|
|
| chr17_+_8175998 | 0.05 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr8_+_31872992 | 0.05 |
ENSDART00000146933
|
c7a
|
complement component 7a |
| chr3_-_31019715 | 0.05 |
ENSDART00000156687
ENSDART00000156127 ENSDART00000153945 |
si:dkeyp-71f10.5
|
si:dkeyp-71f10.5 |
| chr10_+_40583930 | 0.05 |
ENSDART00000134523
|
taar18j
|
trace amine associated receptor 18j |
| chr18_+_33340868 | 0.04 |
ENSDART00000099139
|
v2rh14
|
vomeronasal 2 receptor, h14 |
| chr10_+_21677058 | 0.04 |
ENSDART00000171499
ENSDART00000157516 |
pcdh1gb2
|
protocadherin 1 gamma b 2 |
| chr6_-_36182115 | 0.04 |
ENSDART00000154639
|
brinp3a.2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 2 |
| chr17_+_44780166 | 0.04 |
ENSDART00000156260
|
tmem63c
|
transmembrane protein 63C |
| chr4_-_28958601 | 0.04 |
ENSDART00000111294
|
zgc:174315
|
zgc:174315 |
| chr15_+_6109861 | 0.04 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr7_+_26138240 | 0.04 |
ENSDART00000193750
ENSDART00000184942 |
nat16
|
N-acetyltransferase 16 |
| chr25_-_13320986 | 0.04 |
ENSDART00000169238
|
si:ch211-194m7.8
|
si:ch211-194m7.8 |
| chr1_-_58766424 | 0.04 |
ENSDART00000191010
|
CABZ01096573.1
|
|
| chr7_+_34305903 | 0.04 |
ENSDART00000173575
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr12_+_4971515 | 0.04 |
ENSDART00000161076
|
arhgap27
|
Rho GTPase activating protein 27 |
| chr3_+_21669545 | 0.04 |
ENSDART00000156527
|
crhr1
|
corticotropin releasing hormone receptor 1 |
| chr8_-_4586696 | 0.04 |
ENSDART00000143511
ENSDART00000134324 |
gp1bb
|
glycoprotein Ib (platelet), beta polypeptide |
| chr9_+_29585943 | 0.04 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr2_-_43850637 | 0.04 |
ENSDART00000136077
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr12_+_18524953 | 0.04 |
ENSDART00000090332
|
neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr14_-_6943934 | 0.04 |
ENSDART00000126279
|
clk4a
|
CDC-like kinase 4a |
| chr16_-_17300030 | 0.04 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr13_+_30169681 | 0.04 |
ENSDART00000138326
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr15_+_404891 | 0.04 |
ENSDART00000155682
|
nipsnap2
|
nipsnap homolog 2 |
| chr4_+_69619 | 0.04 |
ENSDART00000164425
|
mansc1
|
MANSC domain containing 1 |
| chr18_+_33550667 | 0.04 |
ENSDART00000141246
ENSDART00000136070 |
si:dkey-47k20.1
|
si:dkey-47k20.1 |
| chr7_-_52417060 | 0.03 |
ENSDART00000148579
|
myzap
|
myocardial zonula adherens protein |
| chr25_-_32464149 | 0.03 |
ENSDART00000152787
|
atp8b4
|
ATPase phospholipid transporting 8B4 |
| chr10_-_22506044 | 0.03 |
ENSDART00000100497
|
zgc:172339
|
zgc:172339 |
| chr3_-_7940522 | 0.03 |
ENSDART00000159710
ENSDART00000167201 |
trim35-25
|
tripartite motif containing 35-25 |
| chr24_-_39121900 | 0.03 |
ENSDART00000020282
|
CU929145.1
|
|
| chr21_-_22648007 | 0.03 |
ENSDART00000121788
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr5_+_38200807 | 0.03 |
ENSDART00000100769
|
hsd20b2
|
hydroxysteroid (20-beta) dehydrogenase 2 |
| chr24_+_7153628 | 0.03 |
ENSDART00000147715
|
si:dkeyp-101e12.1
|
si:dkeyp-101e12.1 |
| chr6_-_49078702 | 0.03 |
ENSDART00000135185
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr7_-_5191797 | 0.03 |
ENSDART00000172955
|
si:ch73-223f5.1
|
si:ch73-223f5.1 |
| chr15_-_33925851 | 0.03 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr18_+_20566817 | 0.02 |
ENSDART00000100716
|
bida
|
BH3 interacting domain death agonist |
| chr25_+_13321307 | 0.02 |
ENSDART00000161284
|
si:ch211-194m7.5
|
si:ch211-194m7.5 |
| chr23_+_32406009 | 0.02 |
ENSDART00000155793
|
si:ch211-66i15.5
|
si:ch211-66i15.5 |
| chr8_+_28900689 | 0.02 |
ENSDART00000141634
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr20_-_31018569 | 0.02 |
ENSDART00000136039
|
fndc1
|
fibronectin type III domain containing 1 |
| chr18_+_45228691 | 0.02 |
ENSDART00000127953
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr22_+_2680895 | 0.02 |
ENSDART00000082245
|
zgc:194221
|
zgc:194221 |
| chr2_-_35103069 | 0.02 |
ENSDART00000111730
|
pappa2
|
pappalysin 2 |
| chr13_+_33651416 | 0.02 |
ENSDART00000180221
|
BX005372.1
|
|
| chr10_+_21584195 | 0.02 |
ENSDART00000100599
|
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr7_+_9922607 | 0.02 |
ENSDART00000184532
ENSDART00000113396 |
cers3a
|
ceramide synthase 3a |
| chr5_+_18925367 | 0.02 |
ENSDART00000051621
|
pgam5
|
PGAM family member 5, serine/threonine protein phosphatase, mitochondrial |
| chr7_-_50883433 | 0.02 |
ENSDART00000174314
|
PAQR9
|
progestin and adipoQ receptor family member 9 |
| chr25_+_12849609 | 0.02 |
ENSDART00000168144
|
ccl33.2
|
chemokine (C-C motif) ligand 33, duplicate 2 |
| chr20_-_1439256 | 0.02 |
ENSDART00000002928
|
themis
|
thymocyte selection associated |
| chr3_-_11373435 | 0.02 |
ENSDART00000192654
|
AL935044.1
|
|
| chr16_-_10547781 | 0.02 |
ENSDART00000166495
|
g6fl
|
g6f-like |
| chr23_+_25856541 | 0.02 |
ENSDART00000145426
ENSDART00000028236 |
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr18_-_32709297 | 0.02 |
ENSDART00000163311
ENSDART00000158288 |
olfcg8
|
olfactory receptor C family, g8 |
| chr5_-_7829657 | 0.02 |
ENSDART00000158374
|
pdlim5a
|
PDZ and LIM domain 5a |
| chr15_+_29088426 | 0.02 |
ENSDART00000187290
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr1_+_49510209 | 0.02 |
ENSDART00000052998
|
si:dkeyp-80c12.4
|
si:dkeyp-80c12.4 |
| chr3_-_58205045 | 0.02 |
ENSDART00000130740
|
si:ch211-256e16.11
|
si:ch211-256e16.11 |
| chr2_+_47927026 | 0.01 |
ENSDART00000143023
|
ftr25
|
finTRIM family, member 25 |
| chr1_+_2025085 | 0.01 |
ENSDART00000008332
|
slc5a3a
|
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3a |
| chr4_-_11106856 | 0.01 |
ENSDART00000150764
|
BX784029.3
|
|
| chr20_+_27194833 | 0.01 |
ENSDART00000150072
|
si:dkey-85n7.8
|
si:dkey-85n7.8 |
| chr15_-_14248047 | 0.01 |
ENSDART00000164762
|
si:ch211-11n16.3
|
si:ch211-11n16.3 |
| chr1_-_26023678 | 0.01 |
ENSDART00000054202
|
si:ch211-145b13.5
|
si:ch211-145b13.5 |
| chr21_+_34814444 | 0.01 |
ENSDART00000161816
|
wdr55
|
WD repeat domain 55 |
| chr4_+_37936249 | 0.01 |
ENSDART00000184022
|
si:dkeyp-82b4.2
|
si:dkeyp-82b4.2 |
| chr15_-_37719015 | 0.01 |
ENSDART00000154875
|
si:dkey-117a8.1
|
si:dkey-117a8.1 |
| chr5_+_9246458 | 0.01 |
ENSDART00000081772
|
susd1
|
sushi domain containing 1 |
| chr8_-_45729248 | 0.01 |
ENSDART00000187736
|
BX088696.1
|
|
| chr4_-_75681004 | 0.01 |
ENSDART00000186296
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr13_-_23031803 | 0.01 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr6_-_32349153 | 0.01 |
ENSDART00000140004
|
angptl3
|
angiopoietin-like 3 |
| chr12_-_23658888 | 0.01 |
ENSDART00000088319
|
map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr10_+_40633990 | 0.01 |
ENSDART00000190489
ENSDART00000139474 |
si:ch211-238p8.31
|
si:ch211-238p8.31 |
| chr22_+_8551813 | 0.01 |
ENSDART00000129690
|
zmp:0000000984
|
zmp:0000000984 |
| chr24_-_39683582 | 0.01 |
ENSDART00000056386
|
CT573356.2
|
|
| chr4_+_59245846 | 0.01 |
ENSDART00000104921
|
si:dkey-105i14.1
|
si:dkey-105i14.1 |
| chr1_+_58400409 | 0.01 |
ENSDART00000111526
|
si:ch73-59p9.2
|
si:ch73-59p9.2 |
| chr5_+_9246018 | 0.01 |
ENSDART00000081769
ENSDART00000183189 |
susd1
|
sushi domain containing 1 |
| chr1_+_26622214 | 0.01 |
ENSDART00000182040
ENSDART00000152643 |
coro2a
|
coronin, actin binding protein, 2A |
| chr22_+_6254194 | 0.01 |
ENSDART00000112388
ENSDART00000135176 |
rnasel4
|
ribonuclease like 4 |
| chr14_+_9462726 | 0.01 |
ENSDART00000063559
|
rell2
|
RELT-like 2 |
| chr17_-_50301313 | 0.01 |
ENSDART00000125772
|
si:ch73-50f9.4
|
si:ch73-50f9.4 |
| chr15_+_17621375 | 0.01 |
ENSDART00000158352
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr2_+_49713592 | 0.01 |
ENSDART00000189624
|
BX323861.3
|
|
| chr25_-_16742438 | 0.01 |
ENSDART00000156091
|
galnt8a.1
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 1 |
| chr12_+_34732262 | 0.00 |
ENSDART00000169634
|
si:dkey-21c1.8
|
si:dkey-21c1.8 |
| chr5_+_19165949 | 0.00 |
ENSDART00000140710
|
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr5_-_37209481 | 0.00 |
ENSDART00000158614
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr15_+_35253247 | 0.00 |
ENSDART00000066737
|
zgc:158328
|
zgc:158328 |
| chr23_+_12072980 | 0.00 |
ENSDART00000141646
|
edn3b
|
endothelin 3b |
| chr25_-_26018240 | 0.00 |
ENSDART00000150800
|
acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr2_-_43850897 | 0.00 |
ENSDART00000005449
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr25_-_32311048 | 0.00 |
ENSDART00000181806
ENSDART00000086334 |
CU372926.1
|
|
| chr20_+_35445462 | 0.00 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr13_+_42011287 | 0.00 |
ENSDART00000131147
|
cyp1b1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr22_+_7923713 | 0.00 |
ENSDART00000167210
|
CABZ01034691.1
|
|
| chr2_+_38742338 | 0.00 |
ENSDART00000177290
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr6_+_41038757 | 0.00 |
ENSDART00000011245
|
entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr7_-_3826639 | 0.00 |
ENSDART00000060330
|
si:dkey-88n24.10
|
si:dkey-88n24.10 |
| chr11_-_45185792 | 0.00 |
ENSDART00000171328
|
si:dkey-93h22.7
|
si:dkey-93h22.7 |
| chr19_-_35733401 | 0.00 |
ENSDART00000066712
|
trappc3
|
trafficking protein particle complex 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CABZ01066694.1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.3 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.1 | 0.5 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.2 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.1 | 0.2 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.3 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.8 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.5 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.1 | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 0.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.4 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.4 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.3 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.2 | GO:0004788 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.0 | 0.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0051379 | epinephrine binding(GO:0051379) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0070840 | dynein complex binding(GO:0070840) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |