Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
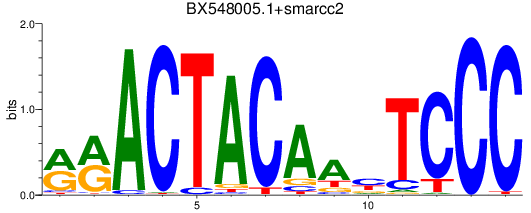
Results for BX548005.1+smarcc2
Z-value: 2.72
Transcription factors associated with BX548005.1+smarcc2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
smarcc2
|
ENSDARG00000077946 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
|
BX548005.1
|
ENSDARG00000110907 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| smarcc2 | dr11_v1_chr6_+_39836474_39836474 | -0.42 | 7.9e-02 | Click! |
Activity profile of BX548005.1+smarcc2 motif
Sorted Z-values of BX548005.1+smarcc2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_29537756 | 12.28 |
ENSDART00000103388
|
wu:fi42e03
|
wu:fi42e03 |
| chr23_+_13528550 | 11.30 |
ENSDART00000099903
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr11_-_16021424 | 9.14 |
ENSDART00000193291
ENSDART00000170731 ENSDART00000104107 |
zgc:173544
|
zgc:173544 |
| chr7_+_41887429 | 7.76 |
ENSDART00000115090
|
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr5_-_43859148 | 7.42 |
ENSDART00000162746
ENSDART00000128763 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr5_+_3891485 | 7.34 |
ENSDART00000129329
ENSDART00000091711 |
rpain
|
RPA interacting protein |
| chr16_-_34285106 | 7.19 |
ENSDART00000044235
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr8_-_36618073 | 6.83 |
ENSDART00000047912
|
gpkow
|
G patch domain and KOW motifs |
| chr6_+_59176470 | 6.78 |
ENSDART00000161720
|
shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr5_-_23675222 | 6.66 |
ENSDART00000135153
|
TBC1D8B
|
si:dkey-110k5.6 |
| chr12_-_13730501 | 6.29 |
ENSDART00000152370
|
foxh1
|
forkhead box H1 |
| chr3_-_26184018 | 6.14 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr20_+_14789305 | 6.08 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr24_-_9989634 | 5.84 |
ENSDART00000115275
|
zgc:152652
|
zgc:152652 |
| chr20_-_28842524 | 5.81 |
ENSDART00000046035
ENSDART00000139843 ENSDART00000129858 ENSDART00000137425 ENSDART00000135720 |
max
|
myc associated factor X |
| chr20_+_14789148 | 5.79 |
ENSDART00000164761
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr24_-_5932982 | 5.70 |
ENSDART00000138412
ENSDART00000135124 ENSDART00000007373 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr8_+_26410197 | 5.58 |
ENSDART00000145836
ENSDART00000053447 |
ifrd2
|
interferon-related developmental regulator 2 |
| chr17_-_16324565 | 5.35 |
ENSDART00000030835
|
hmbox1a
|
homeobox containing 1a |
| chr2_-_57110477 | 5.33 |
ENSDART00000181132
|
slc25a42
|
solute carrier family 25, member 42 |
| chr3_-_26183699 | 5.16 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr23_-_4019928 | 5.13 |
ENSDART00000021062
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr23_-_4019699 | 5.04 |
ENSDART00000159780
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr22_+_25088999 | 5.00 |
ENSDART00000158225
|
rrbp1b
|
ribosome binding protein 1b |
| chr15_-_25435085 | 4.93 |
ENSDART00000112079
|
tlcd2
|
TLC domain containing 2 |
| chr13_-_15929402 | 4.92 |
ENSDART00000090273
|
ttl
|
tubulin tyrosine ligase |
| chr15_-_31147301 | 4.91 |
ENSDART00000157145
ENSDART00000155473 ENSDART00000048103 |
ksr1b
|
kinase suppressor of ras 1b |
| chr11_+_37638873 | 4.84 |
ENSDART00000186384
ENSDART00000184291 ENSDART00000131782 ENSDART00000140502 |
sh2d5
|
SH2 domain containing 5 |
| chr5_-_10082244 | 4.75 |
ENSDART00000036421
|
chek2
|
checkpoint kinase 2 |
| chr18_+_46162204 | 4.73 |
ENSDART00000113545
ENSDART00000147556 |
zgc:113340
|
zgc:113340 |
| chr3_+_27664864 | 4.72 |
ENSDART00000126533
ENSDART00000180848 |
clcn7
|
chloride channel 7 |
| chr16_-_39267185 | 4.71 |
ENSDART00000058550
ENSDART00000133642 |
gpd1l
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr17_+_53186347 | 4.68 |
ENSDART00000155981
|
dph6
|
diphthamine biosynthesis 6 |
| chr8_+_26410539 | 4.62 |
ENSDART00000168780
|
ifrd2
|
interferon-related developmental regulator 2 |
| chr3_+_27665160 | 4.60 |
ENSDART00000103660
|
clcn7
|
chloride channel 7 |
| chr3_+_24060454 | 4.57 |
ENSDART00000143088
|
cbx1a
|
chromobox homolog 1a (HP1 beta homolog Drosophila) |
| chr13_-_33654931 | 4.56 |
ENSDART00000020350
|
snx5
|
sorting nexin 5 |
| chr15_+_2857556 | 4.56 |
ENSDART00000157758
|
mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr14_-_12106603 | 4.55 |
ENSDART00000054619
|
prps1b
|
phosphoribosyl pyrophosphate synthetase 1B |
| chr6_-_39700965 | 4.48 |
ENSDART00000156645
|
espl1
|
extra spindle pole bodies like 1, separase |
| chr13_-_31389661 | 4.45 |
ENSDART00000134630
|
zdhhc16a
|
zinc finger, DHHC-type containing 16a |
| chr23_-_26227805 | 4.41 |
ENSDART00000158082
|
BX927204.1
|
|
| chr15_-_2857961 | 4.34 |
ENSDART00000033263
|
ankrd49
|
ankyrin repeat domain 49 |
| chr8_+_12951155 | 4.33 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr13_+_2861265 | 4.29 |
ENSDART00000170602
ENSDART00000171687 |
XPO5
|
si:ch211-233m11.2 |
| chr16_+_23495600 | 4.24 |
ENSDART00000021092
|
snx27b
|
sorting nexin family member 27b |
| chr18_-_16953978 | 4.18 |
ENSDART00000100126
|
akip1
|
A kinase (PRKA) interacting protein 1 |
| chr23_-_26228077 | 4.18 |
ENSDART00000162423
|
BX927204.1
|
|
| chr18_+_21951551 | 4.16 |
ENSDART00000146261
|
ranbp10
|
RAN binding protein 10 |
| chr4_+_25912308 | 4.15 |
ENSDART00000167845
ENSDART00000136927 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr11_-_35171768 | 4.09 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr8_-_18613948 | 4.09 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr23_+_33947874 | 4.06 |
ENSDART00000136104
|
si:ch211-148l7.4
|
si:ch211-148l7.4 |
| chr24_-_23716097 | 4.05 |
ENSDART00000084954
ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr4_+_25912654 | 4.02 |
ENSDART00000109508
ENSDART00000134218 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr16_-_2390931 | 3.98 |
ENSDART00000149463
|
hace1
|
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| chr21_+_8427059 | 3.95 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr11_-_35171162 | 3.92 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr17_+_25187670 | 3.85 |
ENSDART00000190873
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr8_-_16725959 | 3.84 |
ENSDART00000183593
|
depdc1a
|
DEP domain containing 1a |
| chr5_+_37379825 | 3.80 |
ENSDART00000171826
|
klhl13
|
kelch-like family member 13 |
| chr9_+_37754845 | 3.72 |
ENSDART00000100592
|
pdia5
|
protein disulfide isomerase family A, member 5 |
| chr7_+_33372680 | 3.69 |
ENSDART00000193436
ENSDART00000099988 |
glceb
|
glucuronic acid epimerase b |
| chr16_-_55259199 | 3.68 |
ENSDART00000161130
|
iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr13_+_32454262 | 3.62 |
ENSDART00000057421
|
rdh14a
|
retinol dehydrogenase 14a |
| chr11_-_18283886 | 3.61 |
ENSDART00000019248
|
stimate
|
STIM activating enhance |
| chr18_-_27316599 | 3.61 |
ENSDART00000028294
|
zgc:56106
|
zgc:56106 |
| chr1_+_9153141 | 3.60 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr23_+_13528053 | 3.58 |
ENSDART00000162217
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr20_-_3997531 | 3.54 |
ENSDART00000092217
|
ttc13
|
tetratricopeptide repeat domain 13 |
| chr3_+_35608385 | 3.53 |
ENSDART00000193219
ENSDART00000132703 |
traf7
|
TNF receptor-associated factor 7 |
| chr1_+_49878000 | 3.51 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr4_-_14954327 | 3.50 |
ENSDART00000182729
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr12_+_8474868 | 3.44 |
ENSDART00000062858
|
adoa
|
2-aminoethanethiol (cysteamine) dioxygenase a |
| chr15_-_37589600 | 3.43 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr8_+_48942470 | 3.41 |
ENSDART00000005464
ENSDART00000132035 |
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr7_-_55633475 | 3.41 |
ENSDART00000149478
|
galns
|
galactosamine (N-acetyl)-6-sulfatase |
| chr15_+_25528290 | 3.41 |
ENSDART00000123143
|
npat
|
nuclear protein, ataxia-telangiectasia locus |
| chr18_+_49969568 | 3.40 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr19_-_12648122 | 3.38 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr14_-_26411918 | 3.37 |
ENSDART00000020582
|
tmed9
|
transmembrane p24 trafficking protein 9 |
| chr15_-_44052927 | 3.33 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr6_+_8652310 | 3.32 |
ENSDART00000105098
|
usp40
|
ubiquitin specific peptidase 40 |
| chr16_-_41535690 | 3.28 |
ENSDART00000102662
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr8_+_26396552 | 3.28 |
ENSDART00000087151
|
amt
|
aminomethyltransferase |
| chr9_-_34945566 | 3.27 |
ENSDART00000131908
ENSDART00000059861 |
dcun1d2a
|
DCN1, defective in cullin neddylation 1, domain containing 2a |
| chr14_+_989733 | 3.26 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr12_-_314899 | 3.25 |
ENSDART00000066579
|
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr20_+_3997684 | 3.24 |
ENSDART00000113184
|
arv1
|
ARV1 homolog, fatty acid homeostasis modulator |
| chr10_+_38663917 | 3.20 |
ENSDART00000170182
|
acat1
|
acetyl-CoA acetyltransferase 1 |
| chr8_+_40275830 | 3.19 |
ENSDART00000164414
|
orai1a
|
ORAI calcium release-activated calcium modulator 1a |
| chr17_+_26803470 | 3.18 |
ENSDART00000023470
|
pgrmc2
|
progesterone receptor membrane component 2 |
| chr2_+_36608387 | 3.16 |
ENSDART00000159541
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr5_-_26795438 | 3.14 |
ENSDART00000146124
|
si:ch211-102c2.7
|
si:ch211-102c2.7 |
| chr13_+_39315881 | 3.14 |
ENSDART00000135999
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr2_+_31942390 | 3.09 |
ENSDART00000138684
ENSDART00000146758 ENSDART00000137921 |
otulinb
|
OTU deubiquitinase with linear linkage specificity b |
| chr19_-_432083 | 3.08 |
ENSDART00000165371
|
dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr13_+_28785814 | 3.08 |
ENSDART00000039028
|
nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr23_+_36306539 | 3.07 |
ENSDART00000053267
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr24_+_37709191 | 3.04 |
ENSDART00000066558
|
decr2
|
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr24_-_36301072 | 3.01 |
ENSDART00000062736
|
coasy
|
CoA synthase |
| chr22_+_22302614 | 2.99 |
ENSDART00000049434
|
scamp4
|
secretory carrier membrane protein 4 |
| chr19_-_23227582 | 2.97 |
ENSDART00000042172
|
grb10a
|
growth factor receptor-bound protein 10a |
| chr3_+_866044 | 2.96 |
ENSDART00000126857
|
FP102786.1
|
|
| chr16_-_39195318 | 2.96 |
ENSDART00000058546
|
ebag9
|
estrogen receptor binding site associated, antigen, 9 |
| chr11_-_44999858 | 2.94 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr22_-_506522 | 2.93 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr10_+_37182626 | 2.90 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr19_-_12648408 | 2.88 |
ENSDART00000103692
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr19_+_26072624 | 2.87 |
ENSDART00000147627
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr25_-_3347418 | 2.85 |
ENSDART00000082385
|
golt1bb
|
golgi transport 1Bb |
| chr20_+_9474841 | 2.84 |
ENSDART00000053847
|
rad51b
|
RAD51 paralog B |
| chr11_+_34921492 | 2.81 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr23_+_43870886 | 2.76 |
ENSDART00000102658
ENSDART00000149088 |
nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr8_+_16726386 | 2.75 |
ENSDART00000144621
|
smim15
|
small integral membrane protein 15 |
| chr4_-_14954029 | 2.71 |
ENSDART00000038642
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr7_-_36358303 | 2.70 |
ENSDART00000130028
|
fto
|
fat mass and obesity associated |
| chr6_-_3998199 | 2.69 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr6_+_36821621 | 2.67 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr10_+_44699734 | 2.66 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr21_+_13327527 | 2.66 |
ENSDART00000114294
|
snrpd3l
|
small nuclear ribonucleoprotein D3 polypeptide, like |
| chr17_-_8692722 | 2.65 |
ENSDART00000148931
ENSDART00000192891 |
ctbp2a
|
C-terminal binding protein 2a |
| chr16_-_30655980 | 2.61 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr12_+_36109071 | 2.58 |
ENSDART00000171268
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr7_+_32897723 | 2.57 |
ENSDART00000146789
ENSDART00000140775 ENSDART00000142049 ENSDART00000145523 ENSDART00000135237 ENSDART00000133584 ENSDART00000140800 ENSDART00000137956 ENSDART00000075263 |
tssc4
|
tumor suppressing subtransferable candidate 4 |
| chr23_+_43668756 | 2.55 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr13_-_33054847 | 2.55 |
ENSDART00000057379
ENSDART00000135955 |
vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr17_-_12764360 | 2.53 |
ENSDART00000003418
|
brms1la
|
breast cancer metastasis-suppressor 1-like a |
| chr9_-_3519717 | 2.52 |
ENSDART00000145043
|
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr7_+_7696665 | 2.50 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr10_+_36037977 | 2.49 |
ENSDART00000164678
|
katnal1
|
katanin p60 subunit A-like 1 |
| chr2_-_22659450 | 2.49 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr16_+_38240027 | 2.48 |
ENSDART00000111081
|
prune
|
prune exopolyphosphatase |
| chr8_-_39984593 | 2.46 |
ENSDART00000140127
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr15_-_6966221 | 2.41 |
ENSDART00000165487
ENSDART00000027657 |
mrps22
|
mitochondrial ribosomal protein S22 |
| chr7_+_29863548 | 2.39 |
ENSDART00000136555
ENSDART00000134068 |
tln2a
|
talin 2a |
| chr25_-_25646806 | 2.39 |
ENSDART00000089066
|
tbc1d2b
|
TBC1 domain family, member 2B |
| chr15_-_34878388 | 2.38 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr6_-_58975010 | 2.38 |
ENSDART00000144911
ENSDART00000144514 |
mars
|
methionyl-tRNA synthetase |
| chr3_+_31058464 | 2.38 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr8_+_48943009 | 2.37 |
ENSDART00000180763
|
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr1_-_59243542 | 2.37 |
ENSDART00000163021
|
mvb12a
|
multivesicular body subunit 12A |
| chr5_+_15495351 | 2.37 |
ENSDART00000111646
ENSDART00000114446 |
suds3
|
SDS3 homolog, SIN3A corepressor complex component |
| chr19_-_3726768 | 2.36 |
ENSDART00000161738
|
smim13
|
small integral membrane protein 13 |
| chr2_+_5841108 | 2.33 |
ENSDART00000136180
|
dis3l2
|
DIS3 like 3'-5' exoribonuclease 2 |
| chr16_+_30604387 | 2.31 |
ENSDART00000058785
|
fam210ab
|
family with sequence similarity 210, member Ab |
| chr16_-_32662947 | 2.31 |
ENSDART00000034109
|
coq3
|
coenzyme Q3 methyltransferase |
| chr8_+_52284075 | 2.31 |
ENSDART00000098439
|
ube2d4
|
ubiquitin-conjugating enzyme E2D 4 (putative) |
| chr19_-_46058963 | 2.30 |
ENSDART00000170409
|
nup153
|
nucleoporin 153 |
| chr21_-_11054605 | 2.30 |
ENSDART00000191378
ENSDART00000084061 |
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr18_-_38244871 | 2.30 |
ENSDART00000076399
|
nat10
|
N-acetyltransferase 10 |
| chr3_+_36646054 | 2.29 |
ENSDART00000170013
ENSDART00000159948 |
gspt1l
|
G1 to S phase transition 1, like |
| chr3_+_26905024 | 2.29 |
ENSDART00000077835
|
dexi
|
Dexi homolog (mouse) |
| chr21_+_43485512 | 2.28 |
ENSDART00000026666
|
atg4a
|
autophagy related 4A, cysteine peptidase |
| chr7_-_17712665 | 2.28 |
ENSDART00000149047
|
men1
|
multiple endocrine neoplasia I |
| chr8_-_31384607 | 2.26 |
ENSDART00000164134
ENSDART00000024872 |
creb3l3l
|
cAMP responsive element binding protein 3-like 3 like |
| chr21_+_3854414 | 2.24 |
ENSDART00000122699
|
miga2
|
mitoguardin 2 |
| chr25_-_31763897 | 2.23 |
ENSDART00000041740
|
ubl7a
|
ubiquitin-like 7a (bone marrow stromal cell-derived) |
| chr5_-_67349916 | 2.21 |
ENSDART00000144092
|
mlxip
|
MLX interacting protein |
| chr10_+_36441124 | 2.21 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr9_-_23242684 | 2.20 |
ENSDART00000053282
ENSDART00000179770 |
ccnt2a
|
cyclin T2a |
| chr14_+_30795559 | 2.18 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr14_-_30876708 | 2.17 |
ENSDART00000147597
|
ubl3b
|
ubiquitin-like 3b |
| chr5_+_8196264 | 2.16 |
ENSDART00000174564
ENSDART00000161261 |
lmbrd2a
|
LMBR1 domain containing 2a |
| chr14_+_30775066 | 2.16 |
ENSDART00000139975
ENSDART00000144921 |
atl3
|
atlastin 3 |
| chr24_-_25788841 | 2.15 |
ENSDART00000132235
|
klhl24b
|
kelch-like family member 24b |
| chr3_-_31115601 | 2.15 |
ENSDART00000139090
|
arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr24_-_38131381 | 2.13 |
ENSDART00000105666
|
crp6
|
C-reactive protein 6 |
| chr10_+_44700103 | 2.13 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr16_+_25137483 | 2.10 |
ENSDART00000155666
|
znf576.1
|
zinc finger protein 576, tandem duplicate 1 |
| chr14_+_30774515 | 2.07 |
ENSDART00000191666
|
atl3
|
atlastin 3 |
| chr25_+_15273370 | 2.05 |
ENSDART00000045659
|
tcp11l1
|
t-complex 11, testis-specific-like 1 |
| chr23_-_36305874 | 2.02 |
ENSDART00000147598
ENSDART00000146986 ENSDART00000086985 ENSDART00000133259 |
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr24_+_29352039 | 2.02 |
ENSDART00000101641
|
prmt6
|
protein arginine methyltransferase 6 |
| chr5_+_61361815 | 2.01 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr16_-_34401412 | 2.01 |
ENSDART00000054020
|
hivep3b
|
human immunodeficiency virus type I enhancer binding protein 3b |
| chr23_+_20640484 | 2.01 |
ENSDART00000054691
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr2_+_31308587 | 2.01 |
ENSDART00000027090
|
clul1
|
clusterin-like 1 (retinal) |
| chr23_-_36306337 | 2.00 |
ENSDART00000142760
ENSDART00000136929 ENSDART00000143340 |
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr21_+_170038 | 1.98 |
ENSDART00000157614
|
klhl8
|
kelch-like family member 8 |
| chr3_+_29179329 | 1.97 |
ENSDART00000085216
ENSDART00000190136 |
cacna1i
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr1_+_35494837 | 1.97 |
ENSDART00000140724
|
gab1
|
GRB2-associated binding protein 1 |
| chr18_-_38245062 | 1.96 |
ENSDART00000189092
|
nat10
|
N-acetyltransferase 10 |
| chr14_+_41409697 | 1.95 |
ENSDART00000173335
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr20_-_4049862 | 1.93 |
ENSDART00000158057
|
sprtn
|
SprT-like N-terminal domain |
| chr14_-_30876299 | 1.92 |
ENSDART00000180305
|
ubl3b
|
ubiquitin-like 3b |
| chr16_-_27161410 | 1.91 |
ENSDART00000177503
|
LO017771.1
|
|
| chr8_+_19624589 | 1.85 |
ENSDART00000185698
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr1_+_24557414 | 1.85 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr4_+_15605844 | 1.84 |
ENSDART00000101619
ENSDART00000021384 |
exoc4
|
exocyst complex component 4 |
| chr19_+_9232676 | 1.84 |
ENSDART00000136957
|
kmt2ba
|
lysine (K)-specific methyltransferase 2Ba |
| chr21_-_11054876 | 1.83 |
ENSDART00000146576
|
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr3_+_62140077 | 1.82 |
ENSDART00000108945
|
GID4
|
GID complex subunit 4 homolog |
| chr21_-_30284404 | 1.79 |
ENSDART00000066363
|
zgc:175066
|
zgc:175066 |
| chr9_+_21146862 | 1.77 |
ENSDART00000136365
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr20_-_31743817 | 1.77 |
ENSDART00000137679
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr13_-_26799244 | 1.73 |
ENSDART00000036419
|
vrk2
|
vaccinia related kinase 2 |
| chr23_-_12158685 | 1.72 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr16_+_25107344 | 1.71 |
ENSDART00000033211
|
zgc:66448
|
zgc:66448 |
| chr22_+_8012532 | 1.71 |
ENSDART00000159439
|
CABZ01034698.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of BX548005.1+smarcc2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 2.1 | 6.3 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 1.7 | 13.4 | GO:0044211 | CTP salvage(GO:0044211) |
| 1.6 | 4.8 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 1.6 | 6.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 1.3 | 4.0 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 1.3 | 4.0 | GO:0042245 | RNA repair(GO:0042245) |
| 1.2 | 4.9 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 1.2 | 4.7 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 1.1 | 4.5 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 1.1 | 3.4 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 1.0 | 7.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.0 | 2.9 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 1.0 | 5.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.8 | 5.8 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.8 | 3.3 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.8 | 2.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.8 | 4.7 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.7 | 4.5 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.7 | 2.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.7 | 3.5 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.6 | 2.6 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.6 | 1.9 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.6 | 4.6 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.5 | 3.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.5 | 3.6 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.5 | 4.1 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.5 | 3.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.5 | 2.0 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.5 | 2.3 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.5 | 2.8 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.5 | 3.2 | GO:0032366 | intracellular sterol transport(GO:0032366) |
| 0.5 | 2.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.5 | 3.2 | GO:0030728 | ovulation(GO:0030728) |
| 0.4 | 2.2 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.4 | 7.4 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.4 | 2.7 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.4 | 3.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.4 | 2.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.4 | 1.5 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.4 | 4.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 1.0 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.3 | 2.9 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.3 | 3.3 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.3 | 4.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.3 | 2.8 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.3 | 2.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 3.7 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.3 | 2.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.3 | 1.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.3 | 1.5 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.3 | 3.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.3 | 4.3 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.3 | 2.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 1.1 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.3 | 1.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.3 | 4.1 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.3 | 3.3 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.3 | 1.0 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.3 | 1.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 1.2 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 1.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.2 | 0.9 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.2 | 2.5 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 3.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 3.0 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.2 | 3.0 | GO:0045116 | protein neddylation(GO:0045116) positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.2 | 1.4 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 5.8 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.2 | 3.6 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.2 | 4.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.2 | 4.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 0.9 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 4.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.2 | 4.5 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.2 | 0.3 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.2 | 4.4 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.2 | 0.6 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 0.8 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.3 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 8.4 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.1 | 1.8 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 4.3 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 1.5 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 0.5 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.9 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 1.9 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 2.3 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 1.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.7 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 4.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 7.2 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.1 | 2.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 15.9 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.1 | 3.0 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 1.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.9 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.3 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 3.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 2.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 2.2 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 1.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 7.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 4.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 3.2 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 1.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 3.5 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 9.3 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 1.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 2.7 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 3.3 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.1 | 1.6 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.1 | 0.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 3.9 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.1 | 1.0 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 1.7 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.1 | 1.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 2.3 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 3.2 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.3 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.8 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 2.5 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.3 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 1.0 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 3.1 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 1.1 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 1.4 | GO:0034111 | negative regulation of homotypic cell-cell adhesion(GO:0034111) negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.8 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.3 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 3.4 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.7 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.8 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.0 | 2.0 | GO:0090504 | epiboly(GO:0090504) |
| 0.0 | 7.8 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 1.5 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 2.3 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 1.0 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 2.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0060055 | angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 1.3 | GO:0045089 | positive regulation of innate immune response(GO:0045089) |
| 0.0 | 4.7 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.6 | GO:0043491 | protein kinase B signaling(GO:0043491) |
| 0.0 | 2.2 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.3 | GO:1901653 | cellular response to peptide hormone stimulus(GO:0071375) cellular response to peptide(GO:1901653) |
| 0.0 | 8.9 | GO:0098609 | cell-cell adhesion(GO:0098609) |
| 0.0 | 2.8 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 0.7 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.8 | GO:0060026 | convergent extension(GO:0060026) |
| 0.0 | 0.0 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.9 | 1.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.8 | 6.8 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.8 | 4.0 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.8 | 4.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.6 | 3.6 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.6 | 3.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.5 | 6.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.5 | 1.5 | GO:1990745 | EARP complex(GO:1990745) |
| 0.5 | 1.5 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.5 | 2.8 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.5 | 2.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.4 | 3.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.4 | 2.7 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.4 | 2.7 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.4 | 1.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 3.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.3 | 3.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.3 | 7.1 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.3 | 0.9 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.3 | 2.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 2.2 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.2 | 6.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 1.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 4.9 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.2 | 1.1 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.2 | 2.5 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.2 | 2.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 7.3 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 1.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.2 | 6.4 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 3.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 2.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 2.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 2.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 4.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 4.8 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 2.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 4.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 5.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 2.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 4.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 7.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 2.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 6.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 2.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 8.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 0.3 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 2.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 3.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 2.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.7 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 4.6 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 2.6 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.4 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 3.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 12.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 11.9 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 7.4 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 2.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 3.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 3.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 6.2 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 4.3 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 1.8 | 5.3 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 1.7 | 13.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 1.2 | 4.7 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 1.1 | 4.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 1.1 | 3.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 1.0 | 4.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 1.0 | 4.0 | GO:0043734 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 1.0 | 4.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.9 | 10.2 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.9 | 3.7 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.9 | 4.3 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.8 | 3.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.8 | 4.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.6 | 2.3 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.6 | 6.2 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.6 | 1.7 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.5 | 2.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.5 | 9.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.5 | 4.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.5 | 3.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.5 | 4.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.4 | 2.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.4 | 3.9 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.4 | 2.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 2.0 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.4 | 4.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.4 | 1.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.4 | 7.2 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.4 | 3.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.4 | 3.3 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.3 | 6.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 1.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.3 | 1.9 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.3 | 2.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 1.2 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.3 | 1.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.3 | 2.9 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.3 | 2.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.3 | 2.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 3.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 2.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 2.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 2.5 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 1.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 1.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.7 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.2 | 4.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 3.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 5.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 4.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 1.5 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.2 | 1.9 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 4.0 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.2 | 4.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 8.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.2 | 3.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.2 | 2.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 3.0 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.2 | 2.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 4.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.2 | 0.9 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.2 | 0.6 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.2 | 3.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 2.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 3.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 3.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 7.8 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 2.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 3.0 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 4.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 2.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.3 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.1 | 6.3 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 5.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.1 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.7 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 4.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 4.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 2.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 1.8 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 2.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 2.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 9.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.6 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 4.5 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 2.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 2.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 2.5 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 4.6 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 1.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 4.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 6.8 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 7.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 3.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 2.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 8.3 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 7.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 5.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 2.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 3.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 6.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 3.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 6.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 3.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.4 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 1.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 3.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.6 | 6.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.5 | 3.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.4 | 4.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.4 | 4.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.3 | 1.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 3.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 4.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 4.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.3 | 3.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 7.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.2 | 2.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 11.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 4.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 1.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 4.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.2 | 3.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 3.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 11.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 4.8 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 2.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 3.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 3.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 6.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 3.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |