Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
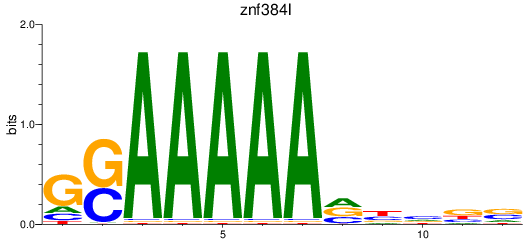
Results for znf384l
Z-value: 1.76
Transcription factors associated with znf384l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf384l
|
ENSDARG00000001015 | zinc finger protein 384 like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf384l | dr11_v1_chr19_-_6983002_6983002 | -0.83 | 8.1e-02 | Click! |
Activity profile of znf384l motif
Sorted Z-values of znf384l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_37829160 | 1.57 |
ENSDART00000099425
|
ctrl
|
chymotrypsin-like |
| chr16_+_35924188 | 1.20 |
ENSDART00000165847
|
sh3d21
|
SH3 domain containing 21 |
| chr12_+_20336070 | 1.19 |
ENSDART00000066385
|
zgc:163057
|
zgc:163057 |
| chr23_+_17865953 | 1.12 |
ENSDART00000014723
ENSDART00000140302 ENSDART00000144800 |
naca
|
nascent polypeptide-associated complex alpha subunit |
| chr16_-_23346095 | 0.93 |
ENSDART00000160546
|
si:dkey-247k7.2
|
si:dkey-247k7.2 |
| chr5_-_6567464 | 0.92 |
ENSDART00000184985
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr12_-_4249000 | 0.91 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr25_+_11456696 | 0.91 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr9_-_33081978 | 0.88 |
ENSDART00000100918
|
zgc:172053
|
zgc:172053 |
| chr3_-_20091964 | 0.85 |
ENSDART00000029386
ENSDART00000020253 ENSDART00000124326 |
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr1_+_1904419 | 0.84 |
ENSDART00000142874
|
si:ch211-132g1.4
|
si:ch211-132g1.4 |
| chr2_+_45191049 | 0.84 |
ENSDART00000165392
|
ccl20a.3
|
chemokine (C-C motif) ligand 20a, duplicate 3 |
| chr23_+_17865554 | 0.83 |
ENSDART00000181009
ENSDART00000162822 |
naca
|
nascent polypeptide-associated complex alpha subunit |
| chr19_+_7930004 | 0.83 |
ENSDART00000160410
|
si:dkey-266f7.4
|
si:dkey-266f7.4 |
| chr14_+_15622817 | 0.81 |
ENSDART00000158624
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr22_-_17671348 | 0.79 |
ENSDART00000137995
|
tjp3
|
tight junction protein 3 |
| chr11_-_20096018 | 0.78 |
ENSDART00000030420
|
ogfrl2
|
opioid growth factor receptor-like 2 |
| chr15_+_22394074 | 0.77 |
ENSDART00000109931
|
oafa
|
OAF homolog a (Drosophila) |
| chr12_+_31713239 | 0.76 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr16_-_21038015 | 0.76 |
ENSDART00000059239
|
snx10b
|
sorting nexin 10b |
| chr16_+_23978978 | 0.74 |
ENSDART00000058964
ENSDART00000135084 |
apoa2
|
apolipoprotein A-II |
| chr4_+_73606482 | 0.73 |
ENSDART00000150765
|
si:ch211-165i18.2
|
si:ch211-165i18.2 |
| chr1_-_59096190 | 0.73 |
ENSDART00000149436
ENSDART00000191532 |
MFAP4 (1 of many)
MFAP4 (1 of many)
|
wu:fj11g02 si:zfos-2330d3.1 |
| chr19_-_5345930 | 0.73 |
ENSDART00000066620
ENSDART00000151398 |
krtt1c19e
|
keratin type 1 c19e |
| chr2_-_45191319 | 0.72 |
ENSDART00000192272
|
CR407590.2
|
|
| chr8_+_21353878 | 0.72 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr13_-_2189761 | 0.72 |
ENSDART00000166255
|
mlip
|
muscular LMNA-interacting protein |
| chr7_+_10610791 | 0.71 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr16_-_11932923 | 0.70 |
ENSDART00000103975
|
cd4-1
|
CD4-1 molecule |
| chr22_-_3344613 | 0.69 |
ENSDART00000165600
|
tbxa2r
|
thromboxane A2 receptor |
| chr6_+_40794015 | 0.69 |
ENSDART00000144479
|
gata2b
|
GATA binding protein 2b |
| chr2_-_44344321 | 0.69 |
ENSDART00000084174
|
lig1
|
ligase I, DNA, ATP-dependent |
| chr22_-_15593824 | 0.68 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr16_-_46664465 | 0.68 |
ENSDART00000135364
|
tmem176l.4
|
transmembrane protein 176l.4 |
| chr18_-_19484919 | 0.67 |
ENSDART00000177690
ENSDART00000184949 |
lctlb
|
lactase-like b |
| chr19_-_13962469 | 0.66 |
ENSDART00000159062
|
paqr7a
|
progestin and adipoQ receptor family member VII, a |
| chr19_-_25519310 | 0.66 |
ENSDART00000089882
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr8_-_25771474 | 0.66 |
ENSDART00000193883
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr21_+_19368720 | 0.65 |
ENSDART00000187759
ENSDART00000185829 ENSDART00000158471 ENSDART00000168728 |
btc
|
betacellulin, epidermal growth factor family member |
| chr3_-_4591643 | 0.64 |
ENSDART00000138144
|
ftr50
|
finTRIM family, member 50 |
| chr5_+_26212621 | 0.64 |
ENSDART00000134432
|
oclnb
|
occludin b |
| chr18_+_26719787 | 0.64 |
ENSDART00000141672
|
alpk3a
|
alpha-kinase 3a |
| chr19_-_25519612 | 0.64 |
ENSDART00000133150
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr25_-_12982193 | 0.64 |
ENSDART00000159617
|
ccl39.5
|
chemokine (C-C motif) ligand 39, duplicate 5 |
| chr1_+_135903 | 0.63 |
ENSDART00000124837
|
f10
|
coagulation factor X |
| chr21_-_11646878 | 0.62 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr1_-_43920371 | 0.62 |
ENSDART00000109283
|
scpp7
|
secretory calcium-binding phosphoprotein 7 |
| chr22_-_10110959 | 0.62 |
ENSDART00000031005
ENSDART00000147580 |
gls2b
|
glutaminase 2b (liver, mitochondrial) |
| chr1_+_40148745 | 0.62 |
ENSDART00000074953
|
zgc:194443
|
zgc:194443 |
| chr22_-_23666504 | 0.61 |
ENSDART00000158665
|
cfh
|
complement factor H |
| chr19_+_7929704 | 0.60 |
ENSDART00000147015
|
si:dkey-266f7.4
|
si:dkey-266f7.4 |
| chr7_+_30051488 | 0.60 |
ENSDART00000169410
|
pstpip1b
|
proline-serine-threonine phosphatase interacting protein 1b |
| chr23_+_36101185 | 0.59 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr1_+_9994811 | 0.59 |
ENSDART00000143719
ENSDART00000110749 |
si:dkeyp-75b4.10
|
si:dkeyp-75b4.10 |
| chr8_+_19356072 | 0.59 |
ENSDART00000063272
|
mpeg1.2
|
macrophage expressed 1, tandem duplicate 2 |
| chr11_-_21404358 | 0.59 |
ENSDART00000129062
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr11_-_42730063 | 0.59 |
ENSDART00000169776
|
si:ch73-106k19.2
|
si:ch73-106k19.2 |
| chr1_+_1838164 | 0.58 |
ENSDART00000006013
|
atp1a1a.5
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 5 |
| chr19_-_977849 | 0.58 |
ENSDART00000172303
|
CABZ01088282.1
|
|
| chr18_+_45645357 | 0.57 |
ENSDART00000010256
|
eif3m
|
eukaryotic translation initiation factor 3, subunit M |
| chr22_-_31059670 | 0.57 |
ENSDART00000022445
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr9_-_45602978 | 0.57 |
ENSDART00000139019
ENSDART00000085763 |
agr1
|
anterior gradient 1 |
| chr7_-_7398350 | 0.55 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr4_+_7677318 | 0.55 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr13_-_33671694 | 0.55 |
ENSDART00000143945
ENSDART00000100504 |
zgc:163030
|
zgc:163030 |
| chr7_-_52417060 | 0.54 |
ENSDART00000148579
|
myzap
|
myocardial zonula adherens protein |
| chr4_+_7817996 | 0.54 |
ENSDART00000166809
|
si:ch1073-67j19.1
|
si:ch1073-67j19.1 |
| chr21_-_22678195 | 0.54 |
ENSDART00000171231
|
gig2g
|
grass carp reovirus (GCRV)-induced gene 2g |
| chr13_-_15994419 | 0.54 |
ENSDART00000079724
ENSDART00000042377 ENSDART00000046079 ENSDART00000050481 ENSDART00000016430 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr21_-_20832482 | 0.53 |
ENSDART00000191928
|
c6
|
complement component 6 |
| chr9_+_8365398 | 0.52 |
ENSDART00000138713
ENSDART00000136847 |
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr22_+_25236888 | 0.52 |
ENSDART00000037286
|
zgc:172218
|
zgc:172218 |
| chr13_-_12646752 | 0.52 |
ENSDART00000042766
|
adh8b
|
alcohol dehydrogenase 8b |
| chr2_-_38287987 | 0.52 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr16_-_30878521 | 0.52 |
ENSDART00000141403
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr10_-_15053507 | 0.52 |
ENSDART00000157446
ENSDART00000170441 |
CLDN23
|
si:ch211-95j8.5 |
| chr13_-_42673978 | 0.52 |
ENSDART00000133848
ENSDART00000099738 ENSDART00000099729 ENSDART00000169083 |
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr8_-_14554785 | 0.51 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr15_+_46356879 | 0.51 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr13_+_46941930 | 0.50 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr18_+_2563756 | 0.50 |
ENSDART00000164147
|
p2ry2.1
|
purinergic receptor P2Y2, tandem duplicate 1 |
| chr5_+_58550291 | 0.50 |
ENSDART00000184983
ENSDART00000044803 |
pou2f3
|
POU class 2 homeobox 3 |
| chr18_-_26894732 | 0.49 |
ENSDART00000147735
ENSDART00000188938 |
BX470164.1
si:dkey-24l11.2
|
si:dkey-24l11.2 |
| chr10_-_17501528 | 0.49 |
ENSDART00000144847
|
slc2a11l
|
solute carrier family 2 (facilitated glucose transporter), member 11-like |
| chr4_+_38550788 | 0.49 |
ENSDART00000157412
|
si:ch211-209n20.1
|
si:ch211-209n20.1 |
| chr10_-_16065185 | 0.49 |
ENSDART00000187266
|
si:dkey-184a18.5
|
si:dkey-184a18.5 |
| chr19_-_17208728 | 0.48 |
ENSDART00000151228
|
stmn1a
|
stathmin 1a |
| chr2_-_42109575 | 0.48 |
ENSDART00000075551
|
adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr12_-_30548244 | 0.48 |
ENSDART00000193616
|
zgc:158404
|
zgc:158404 |
| chr2_+_36112273 | 0.48 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr16_+_33142734 | 0.48 |
ENSDART00000138244
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr25_+_10929790 | 0.48 |
ENSDART00000045182
|
CR339041.2
|
|
| chr8_+_1769475 | 0.47 |
ENSDART00000079073
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr3_-_39488482 | 0.47 |
ENSDART00000135192
|
zgc:100868
|
zgc:100868 |
| chr7_-_38659477 | 0.47 |
ENSDART00000138071
|
npsn
|
nephrosin |
| chr19_+_32856907 | 0.47 |
ENSDART00000148232
|
rpl30
|
ribosomal protein L30 |
| chr5_-_4532516 | 0.46 |
ENSDART00000192398
|
cst14b.1
|
cystatin 14b, tandem duplicate 1 |
| chr14_-_12071447 | 0.46 |
ENSDART00000166116
|
tmsb1
|
thymosin beta 1 |
| chr18_+_35861930 | 0.46 |
ENSDART00000185223
|
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr4_+_12340655 | 0.46 |
ENSDART00000168487
|
pimr214
|
Pim proto-oncogene, serine/threonine kinase, related 214 |
| chr22_+_6674992 | 0.46 |
ENSDART00000144054
|
si:ch211-209l18.2
|
si:ch211-209l18.2 |
| chr11_-_28050559 | 0.46 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr6_-_24143923 | 0.46 |
ENSDART00000157948
|
si:ch73-389b16.1
|
si:ch73-389b16.1 |
| chr22_+_25088999 | 0.46 |
ENSDART00000158225
|
rrbp1b
|
ribosome binding protein 1b |
| chr22_+_25249193 | 0.46 |
ENSDART00000171851
|
si:ch211-226h8.11
|
si:ch211-226h8.11 |
| chr6_+_9651703 | 0.45 |
ENSDART00000122937
|
tcf23
|
transcription factor 23 |
| chr19_+_7043634 | 0.45 |
ENSDART00000133954
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr6_-_27108844 | 0.45 |
ENSDART00000073883
|
dtymk
|
deoxythymidylate kinase (thymidylate kinase) |
| chr15_-_39971756 | 0.45 |
ENSDART00000063789
|
rps5
|
ribosomal protein S5 |
| chr21_-_43616586 | 0.45 |
ENSDART00000036537
|
htr2cl2
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled-like 2 |
| chr16_-_2414063 | 0.45 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
| chr24_+_39990695 | 0.45 |
ENSDART00000040281
|
BX323854.1
|
|
| chr7_-_8577190 | 0.45 |
ENSDART00000173174
|
jac3
|
jacalin 3 |
| chr5_-_19014589 | 0.45 |
ENSDART00000002624
|
ranbp1
|
RAN binding protein 1 |
| chr1_-_59116617 | 0.45 |
ENSDART00000137471
ENSDART00000140490 |
MFAP4 (1 of many)
|
si:zfos-2330d3.7 |
| chr7_-_6345507 | 0.44 |
ENSDART00000173032
|
CU457819.4
|
Histone H3.2 |
| chr25_-_7650335 | 0.44 |
ENSDART00000089034
|
myo5ab
|
myosin VAb |
| chr7_-_48251234 | 0.44 |
ENSDART00000024062
ENSDART00000098904 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr2_+_20406399 | 0.44 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr1_+_41478345 | 0.44 |
ENSDART00000134626
|
dok1a
|
docking protein 1a |
| chr1_+_40140498 | 0.44 |
ENSDART00000101640
|
zgc:152658
|
zgc:152658 |
| chr8_+_41048501 | 0.44 |
ENSDART00000123288
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr24_-_6029314 | 0.44 |
ENSDART00000136155
|
ftr60
|
finTRIM family, member 60 |
| chr23_+_38251864 | 0.43 |
ENSDART00000183498
ENSDART00000129593 |
znf217
|
zinc finger protein 217 |
| chr7_-_20241346 | 0.43 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr23_-_19500559 | 0.43 |
ENSDART00000177414
ENSDART00000145898 |
asb14b
|
ankyrin repeat and SOCS box containing 14b |
| chr9_-_9992697 | 0.43 |
ENSDART00000123415
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr11_-_7261717 | 0.43 |
ENSDART00000128959
|
zgc:113223
|
zgc:113223 |
| chr6_-_52675630 | 0.43 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr16_+_38201840 | 0.43 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr6_-_40446536 | 0.43 |
ENSDART00000153466
|
tatdn2
|
TatD DNase domain containing 2 |
| chr6_-_47506351 | 0.43 |
ENSDART00000128700
|
im:7151449
|
im:7151449 |
| chr13_+_33452096 | 0.43 |
ENSDART00000057370
|
ftr80
|
finTRIM family, member 80 |
| chr19_-_25005609 | 0.42 |
ENSDART00000151129
|
xkr8.2
|
XK, Kell blood group complex subunit-related family, member 8, tandem duplicate 2 |
| chr3_+_24634481 | 0.42 |
ENSDART00000163080
|
si:dkey-68o6.8
|
si:dkey-68o6.8 |
| chr23_+_32044410 | 0.42 |
ENSDART00000048628
|
mylk2
|
myosin light chain kinase 2 |
| chr6_-_46782176 | 0.42 |
ENSDART00000185559
|
igfn1.4
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 4 |
| chr15_+_24563504 | 0.42 |
ENSDART00000140658
ENSDART00000130589 ENSDART00000045549 |
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr2_+_36109002 | 0.42 |
ENSDART00000158978
|
traj28
|
T-cell receptor alpha joining 28 |
| chr17_-_14817458 | 0.42 |
ENSDART00000155907
ENSDART00000156476 |
nid2a
|
nidogen 2a (osteonidogen) |
| chr5_+_23913585 | 0.42 |
ENSDART00000015401
|
ercc6l
|
excision repair cross-complementation group 6-like |
| chr12_+_14084291 | 0.42 |
ENSDART00000189734
|
si:ch211-217a12.1
|
si:ch211-217a12.1 |
| chr23_+_1276006 | 0.42 |
ENSDART00000162294
|
utrn
|
utrophin |
| chr12_-_14143344 | 0.42 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr1_+_29068654 | 0.42 |
ENSDART00000053932
|
cbsa
|
cystathionine-beta-synthase a |
| chr3_+_46635527 | 0.41 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr4_+_12348568 | 0.41 |
ENSDART00000113595
|
pimr170
|
Pim proto-oncogene, serine/threonine kinase, related 170 |
| chr1_+_1689775 | 0.41 |
ENSDART00000048828
|
atp1a1a.4
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 4 |
| chr19_-_5125663 | 0.41 |
ENSDART00000005547
ENSDART00000132642 |
gnb3b
|
guanine nucleotide binding protein (G protein), beta polypeptide 3b |
| chr5_-_68022631 | 0.41 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr3_+_23221047 | 0.41 |
ENSDART00000009393
|
col1a1a
|
collagen, type I, alpha 1a |
| chr16_+_23984179 | 0.41 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr12_+_31608905 | 0.41 |
ENSDART00000152874
ENSDART00000152996 |
CR352342.1
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr7_-_31441420 | 0.41 |
ENSDART00000075398
|
cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr6_+_36821621 | 0.41 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr9_-_33328948 | 0.41 |
ENSDART00000006948
|
rpl8
|
ribosomal protein L8 |
| chr3_+_17689419 | 0.41 |
ENSDART00000192842
ENSDART00000126029 |
dhx58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr17_-_14836320 | 0.41 |
ENSDART00000157051
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr23_-_5818992 | 0.41 |
ENSDART00000148730
|
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr14_-_1280907 | 0.41 |
ENSDART00000186150
|
CABZ01081490.1
|
|
| chr23_-_7674902 | 0.41 |
ENSDART00000185612
ENSDART00000180524 |
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr17_-_8862424 | 0.41 |
ENSDART00000064633
|
nkl.4
|
NK-lysin tandem duplicate 4 |
| chr23_+_28322986 | 0.41 |
ENSDART00000134710
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr12_+_27156943 | 0.41 |
ENSDART00000153030
ENSDART00000001737 |
skap1
|
src kinase associated phosphoprotein 1 |
| chr22_+_1779401 | 0.40 |
ENSDART00000170126
|
znf1154
|
zinc finger protein 1154 |
| chr24_+_15670013 | 0.40 |
ENSDART00000185826
|
CU929414.1
|
|
| chr6_-_29007493 | 0.40 |
ENSDART00000065139
|
gfi1ab
|
growth factor independent 1A transcription repressor b |
| chr13_-_31017960 | 0.40 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr16_-_39570832 | 0.40 |
ENSDART00000039832
|
tgfbr2b
|
transforming growth factor beta receptor 2b |
| chr7_+_21275152 | 0.40 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr20_-_25542183 | 0.40 |
ENSDART00000024350
|
cyp2ad2
|
cytochrome P450, family 2, subfamily AD, polypeptide 2 |
| chr10_-_31015535 | 0.40 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr11_-_25538341 | 0.40 |
ENSDART00000171560
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr10_-_34089779 | 0.40 |
ENSDART00000140070
|
pimr144
|
Pim proto-oncogene, serine/threonine kinase, related 144 |
| chr3_-_3703572 | 0.39 |
ENSDART00000111017
|
si:ch211-163m16.7
|
si:ch211-163m16.7 |
| chr17_+_28103675 | 0.39 |
ENSDART00000188078
|
zgc:91908
|
zgc:91908 |
| chr25_+_10909850 | 0.39 |
ENSDART00000186021
|
CR339041.3
|
|
| chr7_+_35238234 | 0.39 |
ENSDART00000178732
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr7_+_29512673 | 0.39 |
ENSDART00000173895
|
si:dkey-182o15.5
|
si:dkey-182o15.5 |
| chr5_+_8492640 | 0.39 |
ENSDART00000171701
|
osmr
|
oncostatin M receptor |
| chr2_-_58183499 | 0.39 |
ENSDART00000172281
ENSDART00000186262 |
si:ch1073-185p12.2
|
si:ch1073-185p12.2 |
| chr25_+_35502552 | 0.39 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr16_+_35395597 | 0.39 |
ENSDART00000158143
|
si:dkey-34d22.5
|
si:dkey-34d22.5 |
| chr20_-_54014539 | 0.38 |
ENSDART00000060466
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr21_+_15723069 | 0.38 |
ENSDART00000149126
ENSDART00000130628 |
p2rx4a
|
purinergic receptor P2X, ligand-gated ion channel, 4a |
| chr25_-_13319112 | 0.38 |
ENSDART00000179885
|
si:ch211-194m7.8
|
si:ch211-194m7.8 |
| chr1_-_56548178 | 0.38 |
ENSDART00000193361
|
CABZ01059407.1
|
|
| chr8_-_46455874 | 0.38 |
ENSDART00000146985
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr7_-_8374950 | 0.38 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr8_+_8936912 | 0.38 |
ENSDART00000135958
|
si:dkey-83k24.5
|
si:dkey-83k24.5 |
| chr1_-_59102320 | 0.38 |
ENSDART00000193379
|
si:zfos-2330d3.7
|
si:zfos-2330d3.7 |
| chr5_+_29714786 | 0.38 |
ENSDART00000148314
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr7_+_19483277 | 0.38 |
ENSDART00000173750
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr13_+_47810211 | 0.37 |
ENSDART00000122964
|
zmp:0000001006
|
zmp:0000001006 |
| chr13_-_12021566 | 0.37 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr18_+_5273953 | 0.37 |
ENSDART00000165073
|
slc12a1
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr7_-_73717082 | 0.37 |
ENSDART00000164301
ENSDART00000082625 |
BX664721.2
|
|
| chr7_+_34549377 | 0.36 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr22_+_24726066 | 0.36 |
ENSDART00000185534
|
BX539332.2
|
|
| chr2_-_49370042 | 0.36 |
ENSDART00000180515
|
CT583662.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of znf384l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 0.2 | 0.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 0.5 | GO:1903173 | fatty alcohol metabolic process(GO:1903173) |
| 0.2 | 0.5 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.1 | 0.7 | GO:0006266 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.5 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 0.4 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.5 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 1.0 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.4 | GO:0048103 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.1 | 0.4 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.1 | 1.0 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.3 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.3 | GO:0060879 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.1 | 0.5 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.1 | 0.6 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.4 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.1 | 0.2 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.3 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.4 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 0.6 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.3 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.2 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 0.4 | GO:0051299 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) |
| 0.1 | 0.4 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.1 | 0.4 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.3 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.1 | 0.4 | GO:0032648 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.3 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.3 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 1.0 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.3 | GO:0046436 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 0.4 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.1 | 0.3 | GO:0072672 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.1 | 0.4 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.2 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.1 | 1.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.7 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.1 | 0.3 | GO:2000301 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.4 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.3 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.1 | 0.3 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.1 | 0.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0045141 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.2 | GO:0048940 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.0 | 0.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.7 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.3 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.2 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.1 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.2 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.0 | 1.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.2 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.4 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.4 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.3 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.7 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.3 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.0 | 2.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.5 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.6 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 0.3 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0060585 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 1.9 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.3 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.0 | 0.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.4 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.2 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 7.3 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.6 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.8 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.2 | GO:0006388 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.4 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.2 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.2 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.6 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 1.0 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.3 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.3 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 1.6 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.0 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.3 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.0 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.5 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.4 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.0 | 0.1 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.5 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.2 | 0.6 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.1 | 0.6 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.6 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.2 | GO:0030681 | multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 2.3 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.1 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.0 | GO:0070209 | ASTRA complex(GO:0070209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.2 | 0.7 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 0.6 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.2 | 0.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 0.5 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 1.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 0.6 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.2 | 0.5 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.7 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.4 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.4 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 1.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 1.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.3 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 0.3 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 0.3 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.4 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.5 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.1 | 0.4 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.1 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.3 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.3 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 0.7 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.2 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.2 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 0.3 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.1 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.8 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.2 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 1.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.0 | 0.3 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.3 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.4 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.8 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 6.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.8 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.2 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 1.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 1.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.2 | GO:0017171 | serine-type peptidase activity(GO:0008236) serine hydrolase activity(GO:0017171) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.0 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 2.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.9 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.7 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.1 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 2.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |