Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
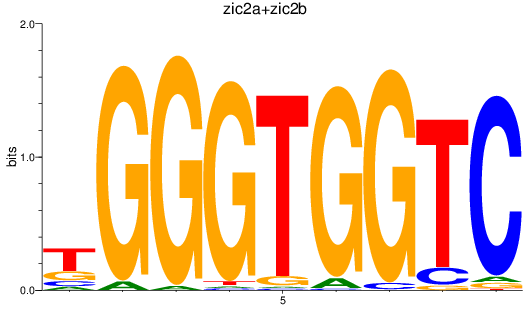
Results for zic2a+zic2b
Z-value: 2.05
Transcription factors associated with zic2a+zic2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zic2a
|
ENSDARG00000015554 | zic family member 2 (odd-paired homolog, Drosophila), a |
|
zic2b
|
ENSDARG00000037178 | zic family member 2 (odd-paired homolog, Drosophila) b |
|
zic2b
|
ENSDARG00000112354 | zic family member 2 (odd-paired homolog, Drosophila) b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zic2a | dr11_v1_chr9_+_31282161_31282161 | 0.88 | 4.9e-02 | Click! |
| zic2b | dr11_v1_chr1_+_29858032_29858032 | 0.79 | 1.1e-01 | Click! |
Activity profile of zic2a+zic2b motif
Sorted Z-values of zic2a+zic2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_83873 | 7.95 |
ENSDART00000162145
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr12_+_47162456 | 2.60 |
ENSDART00000186272
ENSDART00000180811 |
RYR2
|
ryanodine receptor 2 |
| chr19_+_23982466 | 2.53 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr12_+_47162761 | 2.47 |
ENSDART00000192339
ENSDART00000167726 |
RYR2
|
ryanodine receptor 2 |
| chr8_+_31119548 | 2.35 |
ENSDART00000136578
|
syn1
|
synapsin I |
| chr20_-_915234 | 1.82 |
ENSDART00000164816
|
cnr1
|
cannabinoid receptor 1 |
| chr11_+_14147913 | 1.77 |
ENSDART00000022823
ENSDART00000154329 |
plppr3b
|
phospholipid phosphatase related 3b |
| chr3_-_30118856 | 1.70 |
ENSDART00000109953
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr15_+_29085955 | 1.70 |
ENSDART00000156799
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr6_+_36942966 | 1.66 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr11_+_40657612 | 1.63 |
ENSDART00000183271
|
slc45a1
|
solute carrier family 45, member 1 |
| chr11_-_4235811 | 1.57 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr21_-_42055872 | 1.50 |
ENSDART00000144767
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr21_+_17110598 | 1.41 |
ENSDART00000101282
ENSDART00000191864 |
bcr
|
breakpoint cluster region |
| chr3_-_19091024 | 1.38 |
ENSDART00000188485
ENSDART00000110554 |
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr4_+_10365857 | 1.37 |
ENSDART00000138890
|
kcnd2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr13_-_36911118 | 1.32 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr1_+_39597809 | 1.31 |
ENSDART00000122700
|
tenm3
|
teneurin transmembrane protein 3 |
| chr1_+_25783801 | 1.30 |
ENSDART00000102455
|
gucy1a1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr12_+_42436328 | 1.30 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr1_-_45042210 | 1.28 |
ENSDART00000073694
|
smu1b
|
SMU1, DNA replication regulator and spliceosomal factor b |
| chr16_+_29303971 | 1.27 |
ENSDART00000087149
|
hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr14_-_40389699 | 1.24 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr4_+_10366532 | 1.21 |
ENSDART00000189901
|
kcnd2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr19_+_9050852 | 1.20 |
ENSDART00000151031
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr11_+_30162407 | 1.15 |
ENSDART00000190333
ENSDART00000127502 |
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr13_+_3954540 | 1.13 |
ENSDART00000092646
|
lrrc73
|
leucine rich repeat containing 73 |
| chr7_-_45076131 | 1.12 |
ENSDART00000110590
|
zgc:194678
|
zgc:194678 |
| chr17_-_36929332 | 1.10 |
ENSDART00000183454
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr14_+_29609245 | 1.09 |
ENSDART00000043058
|
TENM2
|
si:dkey-34l15.2 |
| chr15_-_26844591 | 1.08 |
ENSDART00000077582
|
pitpnm3
|
PITPNM family member 3 |
| chr3_-_62380146 | 1.08 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr19_-_28360033 | 1.07 |
ENSDART00000186994
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr19_-_26235219 | 1.06 |
ENSDART00000104008
|
dtnbp1b
|
dystrobrevin binding protein 1b |
| chr15_-_12229874 | 1.06 |
ENSDART00000165159
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr10_+_40235959 | 1.03 |
ENSDART00000145862
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr5_-_14564878 | 1.02 |
ENSDART00000160511
|
sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr4_+_4902392 | 1.00 |
ENSDART00000133866
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr24_-_21490628 | 0.99 |
ENSDART00000181546
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr9_-_20372977 | 0.98 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr5_-_43959972 | 0.92 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr9_+_6082793 | 0.92 |
ENSDART00000192045
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr17_-_5769196 | 0.92 |
ENSDART00000113885
|
si:dkey-100n19.2
|
si:dkey-100n19.2 |
| chr11_-_23025949 | 0.90 |
ENSDART00000184859
ENSDART00000167818 ENSDART00000046122 ENSDART00000193120 |
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr9_+_24159725 | 0.87 |
ENSDART00000137756
|
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr14_+_25465346 | 0.85 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr19_-_10196370 | 0.85 |
ENSDART00000091707
|
dbpa
|
D site albumin promoter binding protein a |
| chr5_+_19320554 | 0.84 |
ENSDART00000165119
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr25_+_19955598 | 0.84 |
ENSDART00000091547
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr17_+_23937262 | 0.81 |
ENSDART00000113276
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr6_-_42073367 | 0.81 |
ENSDART00000154304
|
grm2a
|
glutamate receptor, metabotropic 2a |
| chr20_+_26095530 | 0.80 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr14_-_34026316 | 0.80 |
ENSDART00000186062
|
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr9_-_46072805 | 0.77 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr21_-_45588720 | 0.75 |
ENSDART00000186642
ENSDART00000189531 |
LO018363.2
|
|
| chr20_+_1996202 | 0.74 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr2_-_30784502 | 0.73 |
ENSDART00000056735
|
rgs20
|
regulator of G protein signaling 20 |
| chr18_+_9641210 | 0.72 |
ENSDART00000182024
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr18_-_40684756 | 0.72 |
ENSDART00000113799
ENSDART00000139042 |
si:ch211-132b12.7
|
si:ch211-132b12.7 |
| chr2_-_57707039 | 0.71 |
ENSDART00000097685
|
CABZ01060891.1
|
|
| chr3_+_32443395 | 0.70 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr2_-_12242695 | 0.69 |
ENSDART00000158175
|
gpr158b
|
G protein-coupled receptor 158b |
| chr22_-_24858042 | 0.69 |
ENSDART00000137998
ENSDART00000078216 ENSDART00000138378 |
vtg7
|
vitellogenin 7 |
| chr4_-_2219705 | 0.67 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr5_+_52844681 | 0.67 |
ENSDART00000162459
ENSDART00000184914 |
scarb2a
|
scavenger receptor class B, member 2a |
| chr11_+_41560792 | 0.65 |
ENSDART00000127292
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr22_+_28818291 | 0.64 |
ENSDART00000136032
|
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr2_-_30784198 | 0.64 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr16_+_32559821 | 0.63 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr3_-_37351225 | 0.63 |
ENSDART00000174685
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr14_+_24935131 | 0.62 |
ENSDART00000170871
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr20_-_7000225 | 0.62 |
ENSDART00000100098
|
adcy1a
|
adenylate cyclase 1a |
| chr23_+_31000243 | 0.59 |
ENSDART00000085263
|
selenoi
|
selenoprotein I |
| chr6_+_60055168 | 0.59 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr18_+_9637744 | 0.59 |
ENSDART00000190171
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr3_-_25814097 | 0.58 |
ENSDART00000169706
|
ntn1b
|
netrin 1b |
| chr23_+_20422661 | 0.57 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr16_-_2558653 | 0.55 |
ENSDART00000110365
|
adcy3a
|
adenylate cyclase 3a |
| chr11_-_41130239 | 0.55 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr10_+_2876020 | 0.55 |
ENSDART00000136618
ENSDART00000133128 ENSDART00000140803 ENSDART00000141505 |
ccar2
|
cell cycle and apoptosis regulator 2 |
| chr2_+_26656283 | 0.55 |
ENSDART00000133202
ENSDART00000099208 |
asph
|
aspartate beta-hydroxylase |
| chr5_-_30151815 | 0.53 |
ENSDART00000156048
|
zbtb44
|
zinc finger and BTB domain containing 44 |
| chr5_-_58112032 | 0.53 |
ENSDART00000016418
|
drd2b
|
dopamine receptor D2b |
| chr19_-_46566430 | 0.52 |
ENSDART00000166668
|
ext1b
|
exostosin glycosyltransferase 1b |
| chr4_+_6840223 | 0.51 |
ENSDART00000161329
|
dock4b
|
dedicator of cytokinesis 4b |
| chr9_+_54417141 | 0.50 |
ENSDART00000056810
|
drd1b
|
dopamine receptor D1b |
| chr22_-_37611681 | 0.48 |
ENSDART00000028085
|
ttc14
|
tetratricopeptide repeat domain 14 |
| chr18_-_29972709 | 0.48 |
ENSDART00000131207
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr3_+_33745014 | 0.47 |
ENSDART00000159966
|
nacc1a
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing a |
| chr14_-_14705750 | 0.46 |
ENSDART00000168092
|
ogt.1
|
O-linked N-acetylglucosamine (GlcNAc) transferase, tandem duplicate 1 |
| chr22_+_38173960 | 0.46 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr7_-_22632938 | 0.46 |
ENSDART00000159867
ENSDART00000165706 |
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr2_-_33993533 | 0.45 |
ENSDART00000140910
ENSDART00000077304 |
ptch2
|
patched 2 |
| chr14_-_14705335 | 0.44 |
ENSDART00000157392
ENSDART00000159456 |
ogt.1
|
O-linked N-acetylglucosamine (GlcNAc) transferase, tandem duplicate 1 |
| chr9_-_128036 | 0.44 |
ENSDART00000165960
|
FQ377903.2
|
|
| chr2_+_3516913 | 0.43 |
ENSDART00000109346
|
CU693445.1
|
|
| chr24_+_3972260 | 0.43 |
ENSDART00000131753
|
pfkpa
|
phosphofructokinase, platelet a |
| chr6_+_7553085 | 0.42 |
ENSDART00000150939
ENSDART00000151114 |
myh10
|
myosin, heavy chain 10, non-muscle |
| chr18_+_17827149 | 0.42 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr17_-_1407593 | 0.42 |
ENSDART00000157622
ENSDART00000159458 |
zbtb42
|
zinc finger and BTB domain containing 42 |
| chr5_+_13521081 | 0.42 |
ENSDART00000171975
|
si:ch211-230g14.6
|
si:ch211-230g14.6 |
| chr8_+_26226044 | 0.42 |
ENSDART00000186503
|
wdr6
|
WD repeat domain 6 |
| chr25_-_28384954 | 0.40 |
ENSDART00000073500
|
ptprz1a
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1a |
| chr10_-_27197044 | 0.40 |
ENSDART00000137928
|
auts2a
|
autism susceptibility candidate 2a |
| chr20_-_25669813 | 0.39 |
ENSDART00000153118
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr25_+_26923193 | 0.39 |
ENSDART00000187364
|
grm8b
|
glutamate receptor, metabotropic 8b |
| chr20_-_36671660 | 0.39 |
ENSDART00000134819
|
slc5a6a
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6a |
| chr21_-_30415524 | 0.37 |
ENSDART00000101036
|
grpel2
|
GrpE-like 2, mitochondrial |
| chr11_+_3006124 | 0.37 |
ENSDART00000126071
|
cpne5b
|
copine Vb |
| chr21_-_5856050 | 0.36 |
ENSDART00000115367
|
CABZ01071020.1
|
|
| chr17_-_7371564 | 0.35 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr5_+_7393346 | 0.35 |
ENSDART00000164238
|
si:ch73-72b7.1
|
si:ch73-72b7.1 |
| chr21_+_39963851 | 0.35 |
ENSDART00000144435
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr7_-_35126374 | 0.35 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr12_-_6905375 | 0.35 |
ENSDART00000152322
|
pcdh15b
|
protocadherin-related 15b |
| chr7_+_13464878 | 0.34 |
ENSDART00000172969
|
dagla
|
diacylglycerol lipase, alpha |
| chr14_-_10371638 | 0.33 |
ENSDART00000003879
|
fam199x
|
family with sequence similarity 199, X-linked |
| chr15_+_816556 | 0.33 |
ENSDART00000046783
|
zgc:113294
|
zgc:113294 |
| chr5_-_48070779 | 0.33 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr22_+_2686673 | 0.32 |
ENSDART00000161580
|
si:dkey-20i20.15
|
si:dkey-20i20.15 |
| chr13_-_30150592 | 0.32 |
ENSDART00000143093
|
sar1ab
|
secretion associated, Ras related GTPase 1Ab |
| chr4_+_38937575 | 0.32 |
ENSDART00000150385
|
znf997
|
zinc finger protein 997 |
| chr14_+_25464681 | 0.31 |
ENSDART00000067500
ENSDART00000187601 |
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr13_+_42124566 | 0.31 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr15_+_26941063 | 0.31 |
ENSDART00000149957
|
bcas3
|
breast carcinoma amplified sequence 3 |
| chr21_-_45073764 | 0.30 |
ENSDART00000181390
ENSDART00000063714 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr1_-_18446161 | 0.30 |
ENSDART00000089442
|
klhl5
|
kelch-like family member 5 |
| chr5_+_17727310 | 0.30 |
ENSDART00000147657
|
fbrsl1
|
fibrosin-like 1 |
| chr22_-_24818066 | 0.30 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr23_+_31979602 | 0.28 |
ENSDART00000140351
|
pan2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr4_-_75175407 | 0.28 |
ENSDART00000180125
|
CABZ01043953.1
|
|
| chr20_-_45060241 | 0.28 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr9_+_14023386 | 0.28 |
ENSDART00000140199
ENSDART00000124267 |
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr6_-_35106425 | 0.28 |
ENSDART00000165139
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr16_+_54263921 | 0.27 |
ENSDART00000002856
|
drd2l
|
dopamine receptor D2 like |
| chr13_+_36958086 | 0.26 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr7_-_55292116 | 0.26 |
ENSDART00000122603
|
rnf166
|
ring finger protein 166 |
| chr15_+_26940569 | 0.25 |
ENSDART00000189636
ENSDART00000077172 |
bcas3
|
breast carcinoma amplified sequence 3 |
| chr12_-_124264 | 0.25 |
ENSDART00000181551
|
FO818699.1
|
|
| chr4_+_45274792 | 0.25 |
ENSDART00000150295
|
znf1138
|
zinc finger protein 1138 |
| chr17_+_27434626 | 0.25 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr1_+_54737353 | 0.24 |
ENSDART00000130675
ENSDART00000162075 |
pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr22_-_20814450 | 0.23 |
ENSDART00000089076
|
dot1l
|
DOT1-like histone H3K79 methyltransferase |
| chr14_-_48961056 | 0.23 |
ENSDART00000124192
|
si:dkeyp-121d2.7
|
si:dkeyp-121d2.7 |
| chr23_+_23119008 | 0.23 |
ENSDART00000132418
|
samd11
|
sterile alpha motif domain containing 11 |
| chr7_+_17716601 | 0.23 |
ENSDART00000173792
ENSDART00000080825 |
rtn3
|
reticulon 3 |
| chrM_+_4993 | 0.22 |
ENSDART00000093600
|
mt-nd2
|
NADH dehydrogenase 2, mitochondrial |
| chr24_-_1356668 | 0.22 |
ENSDART00000188935
|
nrp1a
|
neuropilin 1a |
| chr18_-_17485419 | 0.22 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr9_+_8372167 | 0.22 |
ENSDART00000181945
ENSDART00000146122 |
si:dkey-90l23.1
|
si:dkey-90l23.1 |
| chr12_-_26582607 | 0.22 |
ENSDART00000045186
|
slc16a5a
|
solute carrier family 16 (monocarboxylate transporter), member 5a |
| chr18_+_48576527 | 0.21 |
ENSDART00000167555
|
kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr2_-_3616363 | 0.21 |
ENSDART00000144699
|
pter
|
phosphotriesterase related |
| chr8_-_53926228 | 0.21 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr23_+_43668756 | 0.21 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr23_+_12072980 | 0.21 |
ENSDART00000141646
|
edn3b
|
endothelin 3b |
| chr23_-_14057191 | 0.20 |
ENSDART00000154908
|
adcy6a
|
adenylate cyclase 6a |
| chr18_+_41250938 | 0.19 |
ENSDART00000087566
|
trip12
|
thyroid hormone receptor interactor 12 |
| chr23_-_27501923 | 0.18 |
ENSDART00000188394
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr17_-_24879003 | 0.18 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chrM_+_8894 | 0.18 |
ENSDART00000093611
|
mt-atp8
|
ATP synthase 8, mitochondrial |
| chr10_+_2876378 | 0.17 |
ENSDART00000192083
|
ccar2
|
cell cycle and apoptosis regulator 2 |
| chr23_-_27152866 | 0.17 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr8_-_47800754 | 0.17 |
ENSDART00000115050
ENSDART00000133669 |
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr6_+_27090800 | 0.16 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr3_-_21087959 | 0.15 |
ENSDART00000184418
|
nlk1
|
nemo-like kinase, type 1 |
| chr5_+_20693724 | 0.15 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr17_-_8562586 | 0.15 |
ENSDART00000154257
ENSDART00000157308 |
fzd3b
|
frizzled class receptor 3b |
| chr17_+_5846202 | 0.15 |
ENSDART00000189713
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr8_-_14239023 | 0.15 |
ENSDART00000090371
|
sin3b
|
SIN3 transcription regulator family member B |
| chr25_+_27744293 | 0.14 |
ENSDART00000103519
ENSDART00000149456 |
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr19_+_5146460 | 0.14 |
ENSDART00000150740
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr13_+_31583034 | 0.14 |
ENSDART00000111763
|
six6a
|
SIX homeobox 6a |
| chr5_+_38654215 | 0.13 |
ENSDART00000144473
|
si:dkey-58f10.14
|
si:dkey-58f10.14 |
| chr22_-_24880824 | 0.13 |
ENSDART00000061165
|
vtg2
|
vitellogenin 2 |
| chr3_+_18846488 | 0.12 |
ENSDART00000148133
|
tmem104
|
transmembrane protein 104 |
| chr13_-_32577386 | 0.12 |
ENSDART00000016535
|
kcns3a
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3a |
| chr2_-_21170517 | 0.12 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr20_+_27194833 | 0.12 |
ENSDART00000150072
|
si:dkey-85n7.8
|
si:dkey-85n7.8 |
| chr12_+_27096835 | 0.12 |
ENSDART00000149475
|
ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr8_+_45294767 | 0.12 |
ENSDART00000191527
|
ubap2b
|
ubiquitin associated protein 2b |
| chr5_+_36439405 | 0.11 |
ENSDART00000102973
|
eda
|
ectodysplasin A |
| chr20_-_28642061 | 0.11 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr22_-_38459316 | 0.11 |
ENSDART00000149683
ENSDART00000098461 |
ptk7a
|
protein tyrosine kinase 7a |
| chr22_-_7809904 | 0.11 |
ENSDART00000182269
ENSDART00000097201 ENSDART00000138522 |
si:ch73-44m9.3
si:ch73-44m9.1
|
si:ch73-44m9.3 si:ch73-44m9.1 |
| chr15_-_24178893 | 0.11 |
ENSDART00000077980
|
pipox
|
pipecolic acid oxidase |
| chr2_+_35615547 | 0.09 |
ENSDART00000135843
|
ankrd45
|
ankyrin repeat domain 45 |
| chr6_+_10094061 | 0.09 |
ENSDART00000150998
ENSDART00000162236 |
atp7b
|
ATPase copper transporting beta |
| chr10_+_43188678 | 0.09 |
ENSDART00000012522
|
vcanb
|
versican b |
| chr16_+_48729947 | 0.08 |
ENSDART00000049051
|
brd2b
|
bromodomain containing 2b |
| chr18_+_3530769 | 0.08 |
ENSDART00000163469
|
si:ch73-338a16.3
|
si:ch73-338a16.3 |
| chr20_-_21910043 | 0.07 |
ENSDART00000152290
ENSDART00000109084 |
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr5_-_25621386 | 0.07 |
ENSDART00000051565
|
cyp1d1
|
cytochrome P450, family 1, subfamily D, polypeptide 1 |
| chr23_+_2542158 | 0.06 |
ENSDART00000182197
|
LO017835.1
|
|
| chr3_-_3372259 | 0.06 |
ENSDART00000140482
|
si:dkey-46g23.1
|
si:dkey-46g23.1 |
| chr4_+_57881965 | 0.05 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr20_+_54349333 | 0.05 |
ENSDART00000132168
ENSDART00000147542 |
znf410
|
zinc finger protein 410 |
| chr4_-_52165969 | 0.05 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr23_+_21978816 | 0.05 |
ENSDART00000087110
|
eif4g3b
|
eukaryotic translation initiation factor 4 gamma, 3b |
| chr19_+_43504480 | 0.05 |
ENSDART00000159421
|
BX649384.3
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of zic2a+zic2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.3 | 1.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.3 | 2.2 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.3 | 1.1 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.2 | 0.6 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.2 | 0.8 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.2 | 0.7 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.2 | 1.3 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 0.6 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.1 | 1.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.5 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.9 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.4 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 0.3 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 3.3 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 1.4 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 1.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.5 | GO:0042311 | vasodilation(GO:0042311) |
| 0.1 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.6 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.4 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.2 | GO:0060585 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.6 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 0.8 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.7 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 1.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.6 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 1.0 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.1 | 0.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.1 | 1.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.2 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.3 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.0 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 1.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.4 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 4.0 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 1.0 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 1.4 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.2 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 1.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 1.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0070977 | ossification involved in bone maturation(GO:0043931) bone maturation(GO:0070977) |
| 0.0 | 0.6 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 1.1 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 3.5 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.1 | GO:0046440 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.9 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.9 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 1.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 1.6 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.4 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 1.0 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.5 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.1 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.6 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.0 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.6 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 1.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.2 | 0.7 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 1.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 1.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 1.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 1.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 2.3 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.8 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 6.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.9 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 1.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.4 | 2.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 0.7 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 1.6 | GO:0008515 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 0.5 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.2 | 1.1 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.2 | 0.9 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.5 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.5 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.6 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.8 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.9 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 1.5 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.6 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.4 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.4 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 1.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.5 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 1.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 1.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.8 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 1.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.2 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 0.8 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.1 | 0.4 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.8 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 2.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 1.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 1.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.4 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.6 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 1.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 1.1 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.3 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.1 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 2.8 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 0.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 1.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 3.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |