Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
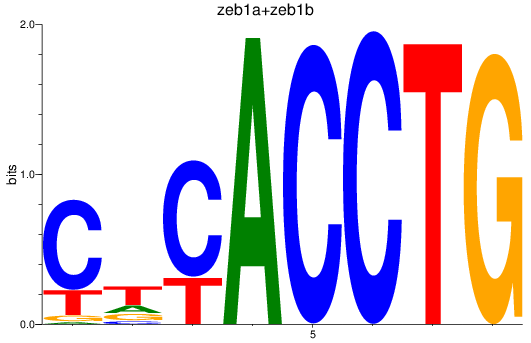
Results for zeb1a+zeb1b
Z-value: 4.85
Transcription factors associated with zeb1a+zeb1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zeb1b
|
ENSDARG00000013207 | zinc finger E-box binding homeobox 1b |
|
zeb1a
|
ENSDARG00000016788 | zinc finger E-box binding homeobox 1a |
|
zeb1b
|
ENSDARG00000113922 | zinc finger E-box binding homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zeb1b | dr11_v1_chr12_-_26851726_26851726 | -0.85 | 7.0e-02 | Click! |
| zeb1a | dr11_v1_chr2_-_43850637_43850637 | -0.75 | 1.5e-01 | Click! |
Activity profile of zeb1a+zeb1b motif
Sorted Z-values of zeb1a+zeb1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_20239141 | 6.92 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr22_+_661711 | 4.88 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr19_-_30404096 | 4.75 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr2_+_68789 | 4.58 |
ENSDART00000058569
|
cldn1
|
claudin 1 |
| chr22_+_7439476 | 4.51 |
ENSDART00000021594
ENSDART00000063389 |
zgc:92041
|
zgc:92041 |
| chr22_+_1170294 | 4.47 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr10_-_2942900 | 4.25 |
ENSDART00000002622
|
oclna
|
occludin a |
| chr6_-_7776612 | 4.20 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr21_-_38153824 | 3.90 |
ENSDART00000151226
|
klf5l
|
Kruppel-like factor 5 like |
| chr19_-_30403922 | 3.87 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr22_+_661505 | 3.78 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr1_-_50247 | 3.47 |
ENSDART00000168428
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr9_+_48007081 | 3.40 |
ENSDART00000060593
ENSDART00000099835 |
zgc:92380
|
zgc:92380 |
| chr16_+_17715243 | 3.39 |
ENSDART00000149437
ENSDART00000149596 |
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr22_-_17677947 | 3.37 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr16_-_26676685 | 3.28 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr25_-_22187397 | 3.26 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr15_-_33964897 | 3.16 |
ENSDART00000172075
ENSDART00000158126 ENSDART00000160456 |
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr19_-_48010490 | 3.10 |
ENSDART00000159938
|
FBXL19
|
zgc:158376 |
| chr19_-_48312109 | 3.04 |
ENSDART00000161103
|
si:ch73-359m17.9
|
si:ch73-359m17.9 |
| chr19_+_791538 | 2.99 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr3_-_61162750 | 2.98 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr2_-_722156 | 2.90 |
ENSDART00000045770
ENSDART00000169498 |
foxq1a
|
forkhead box Q1a |
| chr7_-_53117131 | 2.86 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr8_-_18667693 | 2.82 |
ENSDART00000100516
|
stap2b
|
signal transducing adaptor family member 2b |
| chr16_-_23346095 | 2.77 |
ENSDART00000160546
|
si:dkey-247k7.2
|
si:dkey-247k7.2 |
| chr13_+_33606739 | 2.73 |
ENSDART00000026464
|
cfl1l
|
cofilin 1 (non-muscle), like |
| chr16_+_23947196 | 2.71 |
ENSDART00000103190
ENSDART00000132961 ENSDART00000147690 ENSDART00000142168 |
apoa4b.2
|
apolipoprotein A-IV b, tandem duplicate 2 |
| chr12_-_4243268 | 2.61 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr21_-_25741411 | 2.60 |
ENSDART00000101211
|
cldnh
|
claudin h |
| chr11_+_10984293 | 2.58 |
ENSDART00000065933
|
itgb6
|
integrin, beta 6 |
| chr7_+_35075847 | 2.56 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr7_-_55648336 | 2.50 |
ENSDART00000147792
ENSDART00000135304 ENSDART00000131923 |
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr17_+_2549503 | 2.45 |
ENSDART00000156843
|
si:dkey-248g15.3
|
si:dkey-248g15.3 |
| chr4_-_77557279 | 2.44 |
ENSDART00000180113
|
AL935186.10
|
|
| chr12_-_46228023 | 2.41 |
ENSDART00000153455
|
si:ch211-226h7.6
|
si:ch211-226h7.6 |
| chr22_+_11756040 | 2.39 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr12_-_11349899 | 2.37 |
ENSDART00000079645
|
zgc:174164
|
zgc:174164 |
| chr5_-_65021736 | 2.37 |
ENSDART00000162368
ENSDART00000161876 |
anxa1c
|
annexin A1c |
| chr10_+_2715548 | 2.34 |
ENSDART00000130793
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr12_-_46252062 | 2.33 |
ENSDART00000153223
|
si:ch211-226h7.5
|
si:ch211-226h7.5 |
| chr3_-_33970190 | 2.30 |
ENSDART00000151238
|
ighj2-5
|
ighj2-5 |
| chr22_+_7439186 | 2.28 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr17_-_32426392 | 2.26 |
ENSDART00000148455
ENSDART00000149885 ENSDART00000179314 |
grhl1
|
grainyhead-like transcription factor 1 |
| chr1_+_56180416 | 2.23 |
ENSDART00000089358
|
crb3b
|
crumbs homolog 3b |
| chr15_-_107900 | 2.22 |
ENSDART00000193184
|
apoa1b
|
apolipoprotein A-Ib |
| chr16_-_20312146 | 2.22 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr17_-_2596125 | 2.20 |
ENSDART00000175740
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr2_+_15612755 | 2.19 |
ENSDART00000003035
|
amy2a
|
amylase, alpha 2A (pancreatic) |
| chr25_+_22107643 | 2.18 |
ENSDART00000089680
|
sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr14_+_48862987 | 2.15 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr21_+_30351256 | 2.15 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr22_-_15578402 | 2.14 |
ENSDART00000062986
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr1_+_58922027 | 2.14 |
ENSDART00000159479
|
trip10b
|
thyroid hormone receptor interactor 10b |
| chr22_+_7480465 | 2.14 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr13_+_24750078 | 2.12 |
ENSDART00000021053
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr19_+_10855158 | 2.11 |
ENSDART00000172219
ENSDART00000170826 |
apoea
|
apolipoprotein Ea |
| chr1_-_45177373 | 2.10 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr24_-_20444844 | 2.09 |
ENSDART00000048940
|
vill
|
villin-like |
| chr22_+_3914318 | 2.03 |
ENSDART00000188774
ENSDART00000082034 |
FO904903.1
|
Danio rerio major histocompatibility complex class I ULA (mhc1ula), mRNA. |
| chr5_-_37959874 | 1.97 |
ENSDART00000031719
|
mpzl2b
|
myelin protein zero-like 2b |
| chr19_-_5332784 | 1.97 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr10_-_34002185 | 1.95 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr21_-_22648007 | 1.92 |
ENSDART00000121788
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr3_-_19561058 | 1.91 |
ENSDART00000079323
|
zgc:163079
|
zgc:163079 |
| chr17_+_2552453 | 1.89 |
ENSDART00000189889
|
si:dkey-248g15.3
|
si:dkey-248g15.3 |
| chr12_+_5708400 | 1.89 |
ENSDART00000017191
|
dlx3b
|
distal-less homeobox 3b |
| chr11_-_11518469 | 1.88 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr17_-_26867725 | 1.88 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr23_-_10175898 | 1.86 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr17_-_30652738 | 1.86 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr11_+_37216668 | 1.85 |
ENSDART00000173076
|
zgc:112265
|
zgc:112265 |
| chr22_+_7497319 | 1.83 |
ENSDART00000034564
|
CELA1 (1 of many)
|
zgc:92511 |
| chr25_-_169291 | 1.80 |
ENSDART00000128344
|
lipcb
|
lipase, hepatic b |
| chr1_-_52437056 | 1.78 |
ENSDART00000138337
|
si:ch211-217k17.12
|
si:ch211-217k17.12 |
| chr19_-_40198478 | 1.78 |
ENSDART00000191736
|
grn2
|
granulin 2 |
| chr4_-_68563862 | 1.77 |
ENSDART00000182970
|
BX548011.2
|
|
| chr4_+_76671012 | 1.76 |
ENSDART00000005585
|
ms4a17a.2
|
membrane-spanning 4-domains, subfamily A, member 17a.2 |
| chr16_+_23921777 | 1.75 |
ENSDART00000163213
|
apoa4b.3
|
apolipoprotein A-IV b, tandem duplicate 3 |
| chr21_-_30293224 | 1.74 |
ENSDART00000101051
|
slbp2
|
stem-loop binding protein 2 |
| chr15_-_43164591 | 1.71 |
ENSDART00000171305
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr18_-_46354269 | 1.71 |
ENSDART00000010813
|
foxa3
|
forkhead box A3 |
| chr15_-_41714329 | 1.70 |
ENSDART00000154113
|
CR853294.1
|
|
| chr10_-_35149513 | 1.70 |
ENSDART00000063434
ENSDART00000131291 |
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr1_-_2051599 | 1.69 |
ENSDART00000041936
|
oxgr1a.1
|
oxoglutarate (alpha-ketoglutarate) receptor 1a, tandem duplicate 1 |
| chr1_-_45146834 | 1.68 |
ENSDART00000144997
|
si:ch211-239f4.6
|
si:ch211-239f4.6 |
| chr8_+_2478862 | 1.68 |
ENSDART00000131739
|
si:dkeyp-51b9.3
|
si:dkeyp-51b9.3 |
| chr16_+_23921610 | 1.67 |
ENSDART00000143855
|
apoa4b.3
|
apolipoprotein A-IV b, tandem duplicate 3 |
| chr4_-_68564988 | 1.67 |
ENSDART00000191212
|
BX548011.3
|
|
| chr24_-_31904924 | 1.66 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr18_-_38271298 | 1.65 |
ENSDART00000143016
|
caprin1b
|
cell cycle associated protein 1b |
| chr17_+_26965351 | 1.65 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr25_-_17395315 | 1.64 |
ENSDART00000064596
|
cyp2x8
|
cytochrome P450, family 2, subfamily X, polypeptide 8 |
| chr20_-_3238110 | 1.64 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr20_+_1398564 | 1.63 |
ENSDART00000002242
|
leg1.2
|
liver-enriched gene 1, tandem duplicate 2 |
| chr21_-_25741096 | 1.62 |
ENSDART00000181756
|
cldnh
|
claudin h |
| chr11_-_45429199 | 1.62 |
ENSDART00000173111
|
rfc4
|
replication factor C (activator 1) 4 |
| chr14_+_14568437 | 1.61 |
ENSDART00000164749
|
pcdh20
|
protocadherin 20 |
| chr20_-_42702832 | 1.60 |
ENSDART00000134689
ENSDART00000045816 |
plg
|
plasminogen |
| chr12_-_46315898 | 1.60 |
ENSDART00000153138
|
si:ch211-226h7.3
|
si:ch211-226h7.3 |
| chr3_-_33970998 | 1.60 |
ENSDART00000151527
|
ighj2-2
|
immunoglobulin heavy joining 2-2 |
| chr16_+_17714664 | 1.59 |
ENSDART00000149042
|
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr8_+_25302172 | 1.58 |
ENSDART00000046182
ENSDART00000145316 |
gstm.3
|
glutathione S-transferase mu tandem duplicate 3 |
| chr22_+_7472420 | 1.57 |
ENSDART00000132049
|
CELA1 (1 of many)
|
si:dkey-57c15.4 |
| chr1_+_26411496 | 1.57 |
ENSDART00000112263
|
arhgef38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr7_+_30392613 | 1.57 |
ENSDART00000075508
|
lipca
|
lipase, hepatic a |
| chr21_-_7928101 | 1.56 |
ENSDART00000151543
ENSDART00000114982 |
f2rl1.2
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 2 |
| chr2_+_49457626 | 1.55 |
ENSDART00000129967
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr2_+_49457449 | 1.54 |
ENSDART00000185470
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr20_+_1412193 | 1.54 |
ENSDART00000064419
|
leg1.1
|
liver-enriched gene 1, tandem duplicate 1 |
| chr20_+_305035 | 1.54 |
ENSDART00000104807
|
si:dkey-119m7.4
|
si:dkey-119m7.4 |
| chr5_+_26212621 | 1.53 |
ENSDART00000134432
|
oclnb
|
occludin b |
| chr22_+_7742211 | 1.53 |
ENSDART00000140896
|
CELA1 (1 of many)
|
zgc:92511 |
| chr7_-_34265481 | 1.51 |
ENSDART00000173596
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr7_-_38658411 | 1.51 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr15_-_47929455 | 1.50 |
ENSDART00000064462
|
psma6l
|
proteasome subunit alpha 6, like |
| chr5_+_30384554 | 1.50 |
ENSDART00000135483
|
zgc:158412
|
zgc:158412 |
| chr23_+_31815423 | 1.50 |
ENSDART00000075730
ENSDART00000075726 |
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr24_-_26369185 | 1.49 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr25_+_10416583 | 1.49 |
ENSDART00000073907
|
ehf
|
ets homologous factor |
| chr16_-_42186093 | 1.49 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr4_+_18843015 | 1.47 |
ENSDART00000152086
ENSDART00000066977 ENSDART00000132567 |
bik
|
BCL2 interacting killer |
| chr17_+_24843401 | 1.47 |
ENSDART00000110179
|
cx34.4
|
connexin 34.4 |
| chr8_-_11170114 | 1.46 |
ENSDART00000133532
|
si:ch211-204d2.4
|
si:ch211-204d2.4 |
| chr16_+_28754403 | 1.46 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr13_-_15994419 | 1.45 |
ENSDART00000079724
ENSDART00000042377 ENSDART00000046079 ENSDART00000050481 ENSDART00000016430 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr10_-_15053507 | 1.45 |
ENSDART00000157446
ENSDART00000170441 |
CLDN23
|
si:ch211-95j8.5 |
| chr13_-_36034582 | 1.45 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr8_+_999421 | 1.45 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr3_+_49021079 | 1.45 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr9_-_44948488 | 1.44 |
ENSDART00000059228
|
vil1
|
villin 1 |
| chr23_-_29824146 | 1.44 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr22_+_19494996 | 1.44 |
ENSDART00000140829
ENSDART00000135168 |
si:dkey-78l4.10
|
si:dkey-78l4.10 |
| chr21_-_7940043 | 1.44 |
ENSDART00000099733
ENSDART00000136671 |
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr1_-_7951002 | 1.43 |
ENSDART00000138187
|
si:dkey-79f11.8
|
si:dkey-79f11.8 |
| chr7_-_8416750 | 1.43 |
ENSDART00000181857
|
jac1
|
jacalin 1 |
| chr2_-_32237916 | 1.43 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr4_-_178510 | 1.43 |
ENSDART00000169805
|
eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr19_+_48141623 | 1.43 |
ENSDART00000170439
|
btr30
|
bloodthirsty-related gene family, member 30 |
| chr17_+_43868441 | 1.42 |
ENSDART00000134272
|
zgc:66313
|
zgc:66313 |
| chr5_-_22590739 | 1.41 |
ENSDART00000141555
|
si:dkey-103e21.5
|
si:dkey-103e21.5 |
| chr19_+_4800549 | 1.41 |
ENSDART00000056235
|
st3gal1l
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1, like |
| chr1_-_7603734 | 1.41 |
ENSDART00000009315
|
mxb
|
myxovirus (influenza) resistance B |
| chr7_+_55149001 | 1.40 |
ENSDART00000148642
|
cdh31
|
cadherin 31 |
| chr15_-_108414 | 1.39 |
ENSDART00000170044
|
apoa1b
|
apolipoprotein A-Ib |
| chr23_+_46157638 | 1.38 |
ENSDART00000076048
|
btr32
|
bloodthirsty-related gene family, member 32 |
| chr3_+_8224622 | 1.38 |
ENSDART00000138670
|
si:ch73-379f5.5
|
si:ch73-379f5.5 |
| chr4_-_13518381 | 1.38 |
ENSDART00000067153
|
ifng1-1
|
interferon, gamma 1-1 |
| chr14_+_15543331 | 1.38 |
ENSDART00000167025
|
si:dkey-203a12.7
|
si:dkey-203a12.7 |
| chr21_-_217589 | 1.38 |
ENSDART00000185017
|
CZQB01146713.1
|
|
| chr21_-_3700334 | 1.37 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr14_+_32964 | 1.37 |
ENSDART00000166173
|
lyar
|
Ly1 antibody reactive homolog (mouse) |
| chr13_+_18533005 | 1.36 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr7_+_32901658 | 1.36 |
ENSDART00000115420
|
ano9b
|
anoctamin 9b |
| chr7_+_73670137 | 1.36 |
ENSDART00000050357
|
btr12
|
bloodthirsty-related gene family, member 12 |
| chr3_-_33970431 | 1.35 |
ENSDART00000151313
|
ighj2-4
|
ighj2-4 |
| chr3_+_21189766 | 1.35 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr11_+_329687 | 1.34 |
ENSDART00000172882
|
cyp27b1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr15_-_576135 | 1.34 |
ENSDART00000124170
|
cbln20
|
cerebellin 20 |
| chr17_-_2595736 | 1.34 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr18_+_44703343 | 1.33 |
ENSDART00000131510
|
b3gnt2l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2, like |
| chr1_+_59538755 | 1.33 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr13_-_21672131 | 1.33 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr14_+_6953244 | 1.33 |
ENSDART00000159470
|
rack1
|
receptor for activated C kinase 1 |
| chr20_+_26556174 | 1.32 |
ENSDART00000138492
|
irf4b
|
interferon regulatory factor 4b |
| chr15_-_21165237 | 1.32 |
ENSDART00000157069
|
A2ML1 (1 of many)
|
si:ch211-212c13.8 |
| chr25_-_16600811 | 1.32 |
ENSDART00000011397
|
cpa2
|
carboxypeptidase A2 (pancreatic) |
| chr25_+_3217419 | 1.32 |
ENSDART00000104859
|
rccd1
|
RCC1 domain containing 1 |
| chr6_-_609880 | 1.31 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr3_-_39488482 | 1.31 |
ENSDART00000135192
|
zgc:100868
|
zgc:100868 |
| chr16_-_9830451 | 1.31 |
ENSDART00000148528
|
grhl2a
|
grainyhead-like transcription factor 2a |
| chr6_+_153146 | 1.30 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr2_-_47620806 | 1.30 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr24_-_36593876 | 1.30 |
ENSDART00000160901
|
CABZ01055365.1
|
|
| chr1_-_59095035 | 1.29 |
ENSDART00000130791
|
MFAP4 (1 of many)
|
wu:fj11g02 |
| chr7_+_66565930 | 1.29 |
ENSDART00000154597
|
tmem176l.3b
|
transmembrane protein 176l.3b |
| chr16_-_46664465 | 1.29 |
ENSDART00000135364
|
tmem176l.4
|
transmembrane protein 176l.4 |
| chr3_-_39488639 | 1.27 |
ENSDART00000161644
|
zgc:100868
|
zgc:100868 |
| chr5_-_67471375 | 1.27 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr17_-_42988356 | 1.27 |
ENSDART00000024558
|
zgc:92137
|
zgc:92137 |
| chr6_+_36877968 | 1.26 |
ENSDART00000155187
|
traf3ip2l
|
TRAF3 interacting protein 2-like |
| chr2_-_58075414 | 1.26 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr4_-_12725513 | 1.25 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr14_+_22022441 | 1.25 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr22_+_2960280 | 1.25 |
ENSDART00000134188
ENSDART00000134451 ENSDART00000139894 |
impact
|
impact RWD domain protein |
| chr2_-_8648440 | 1.25 |
ENSDART00000135743
|
si:ch211-71m22.3
|
si:ch211-71m22.3 |
| chr7_+_35068036 | 1.25 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr3_+_29469283 | 1.25 |
ENSDART00000103592
|
fam83fa
|
family with sequence similarity 83, member Fa |
| chr21_+_10577527 | 1.25 |
ENSDART00000165070
ENSDART00000127963 |
ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr6_-_33913184 | 1.25 |
ENSDART00000146373
|
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr23_-_31428763 | 1.24 |
ENSDART00000053545
|
zgc:153284
|
zgc:153284 |
| chr5_-_33236637 | 1.24 |
ENSDART00000085512
ENSDART00000144694 |
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr19_-_15192638 | 1.24 |
ENSDART00000048151
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr12_+_20352400 | 1.23 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr16_+_13427967 | 1.23 |
ENSDART00000038196
|
zgc:101640
|
zgc:101640 |
| chr20_-_17041025 | 1.22 |
ENSDART00000063764
|
si:dkey-5n18.1
|
si:dkey-5n18.1 |
| chr21_-_22676323 | 1.22 |
ENSDART00000167392
|
gig2h
|
grass carp reovirus (GCRV)-induced gene 2h |
| chr10_+_9561066 | 1.22 |
ENSDART00000136281
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr18_-_977075 | 1.21 |
ENSDART00000032392
|
dhdhl
|
dihydrodiol dehydrogenase (dimeric), like |
| chr21_-_25295087 | 1.21 |
ENSDART00000087910
ENSDART00000147860 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
Network of associatons between targets according to the STRING database.
First level regulatory network of zeb1a+zeb1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.4 | GO:0061438 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 1.1 | 5.3 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.9 | 3.7 | GO:0097242 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.9 | 2.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.9 | 8.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.8 | 3.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.6 | 1.9 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.6 | 1.7 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.5 | 1.6 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.5 | 3.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.5 | 2.9 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.5 | 1.4 | GO:0046415 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.4 | 1.3 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.4 | 2.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.4 | 3.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.4 | 2.4 | GO:2000516 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.4 | 2.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.4 | 1.9 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.4 | 1.1 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.4 | 3.0 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.4 | 3.4 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.4 | 1.4 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.4 | 6.1 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.3 | 1.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 1.0 | GO:0000103 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.3 | 1.0 | GO:0030237 | female sex determination(GO:0030237) |
| 0.3 | 0.3 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) RNA 5'-end processing(GO:0000966) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.3 | 0.9 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.3 | 1.7 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 0.8 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.3 | 1.0 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.3 | 1.5 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.3 | 1.3 | GO:0070301 | response to acidic pH(GO:0010447) cellular response to hydrogen peroxide(GO:0070301) |
| 0.2 | 0.7 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 1.5 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.2 | 2.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 11.5 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.2 | 1.1 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 | 1.1 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.2 | 0.2 | GO:0032768 | regulation of monooxygenase activity(GO:0032768) positive regulation of monooxygenase activity(GO:0032770) |
| 0.2 | 1.6 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 1.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 | 0.7 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.2 | 0.7 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.2 | 1.3 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.2 | 0.6 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.2 | 1.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.2 | 0.6 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.2 | 2.0 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.2 | 0.4 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 1.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.2 | 0.6 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.2 | 0.8 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 | 1.2 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.2 | 2.9 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 4.9 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.2 | 0.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 1.8 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.2 | 1.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.2 | 2.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 5.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.2 | 0.5 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.2 | 0.7 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.2 | 0.9 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.2 | 4.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 1.6 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.2 | 3.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.2 | 1.0 | GO:0046874 | quinolinate biosynthetic process(GO:0019805) quinolinate metabolic process(GO:0046874) |
| 0.2 | 1.5 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.2 | 0.7 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.2 | 0.2 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.2 | 1.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.2 | 0.7 | GO:1902745 | positive regulation of lamellipodium organization(GO:1902745) |
| 0.2 | 1.0 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.2 | 0.5 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.2 | 0.7 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.2 | 2.7 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.2 | 0.5 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 0.5 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.2 | 1.5 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.2 | 0.8 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.2 | 0.8 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.2 | 0.5 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.2 | 0.5 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.2 | 4.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 0.9 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 0.5 | GO:0043385 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.2 | 0.6 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.2 | 1.1 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.2 | 0.9 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.2 | 2.6 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 1.4 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 3.0 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.1 | 1.5 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.1 | 0.7 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.4 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.7 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.4 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 3.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.4 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.1 | 0.7 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.1 | 0.4 | GO:0071706 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.1 | 1.0 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.4 | GO:1903173 | fatty alcohol metabolic process(GO:1903173) |
| 0.1 | 0.7 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.7 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.1 | 0.4 | GO:0098801 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) regulation of renal system process(GO:0098801) |
| 0.1 | 0.9 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.9 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 2.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 0.4 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) |
| 0.1 | 0.7 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.3 | GO:0048521 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.5 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) |
| 0.1 | 0.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.8 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.3 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.1 | 0.5 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.1 | 1.2 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.9 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.4 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.1 | 0.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 1.5 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.1 | 1.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.4 | GO:0055057 | neuroblast division(GO:0055057) positive regulation of stem cell proliferation(GO:2000648) |
| 0.1 | 0.7 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.1 | 1.3 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 0.5 | GO:0033273 | response to vitamin(GO:0033273) |
| 0.1 | 1.8 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.1 | 0.8 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 14.1 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.1 | 0.9 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 2.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.7 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.8 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.1 | 0.4 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 2.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.8 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 0.1 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.1 | 0.3 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 1.2 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 1.1 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.4 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 1.4 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 2.0 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.1 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.8 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 0.2 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 0.6 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 2.1 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.5 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.4 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 1.4 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.7 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 1.0 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 0.3 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.3 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 0.4 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.1 | 0.8 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 2.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 1.0 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 2.2 | GO:0043588 | skin development(GO:0043588) |
| 0.1 | 0.7 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.8 | GO:0042664 | negative regulation of endodermal cell fate specification(GO:0042664) |
| 0.1 | 0.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 0.4 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 1.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.7 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.4 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.1 | 0.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.4 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.1 | 0.9 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.1 | 0.3 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.1 | 0.1 | GO:0032366 | intracellular sterol transport(GO:0032366) |
| 0.1 | 0.6 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.8 | GO:0042531 | tyrosine phosphorylation of STAT protein(GO:0007260) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 5.9 | GO:0007492 | endoderm development(GO:0007492) |
| 0.1 | 0.3 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 0.5 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 0.3 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 0.9 | GO:0030719 | P granule organization(GO:0030719) |
| 0.1 | 0.5 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 0.8 | GO:0046632 | alpha-beta T cell differentiation(GO:0046632) |
| 0.1 | 0.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.6 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 1.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 1.6 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.1 | 0.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.8 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.7 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.7 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 1.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.4 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 0.1 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.4 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.6 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.1 | 0.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 1.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 3.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.4 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.5 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.9 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.5 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 0.6 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 0.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.2 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 3.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.4 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 0.5 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.5 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.1 | 0.3 | GO:0090208 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) positive regulation of triglyceride metabolic process(GO:0090208) |
| 0.1 | 1.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.2 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 0.3 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 1.2 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 0.2 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.1 | 0.7 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.2 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 1.0 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 0.2 | GO:0070197 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.9 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.4 | GO:0001840 | neural plate development(GO:0001840) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 1.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 0.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.3 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 0.6 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 1.2 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 0.7 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.5 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.9 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 1.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.7 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.4 | GO:0072048 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) pattern specification involved in pronephros development(GO:0039017) pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.1 | 0.2 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.8 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 0.2 | GO:0046755 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.1 | 0.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.2 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.1 | 0.2 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 7.3 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.1 | 0.8 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 5.2 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.1 | 0.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.4 | GO:0003188 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.1 | 0.2 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 0.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 2.6 | GO:0010950 | positive regulation of endopeptidase activity(GO:0010950) positive regulation of peptidase activity(GO:0010952) |
| 0.0 | 0.2 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 5.8 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.8 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.3 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 1.1 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 3.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0048855 | adenohypophysis morphogenesis(GO:0048855) |
| 0.0 | 1.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.8 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.4 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 5.6 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 1.4 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 4.0 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.2 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.0 | 0.2 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.0 | 0.7 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.3 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 1.9 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.9 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 1.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 1.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.9 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.2 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 1.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.0 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.5 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 1.7 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.3 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 1.1 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.1 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.7 | GO:0002440 | production of molecular mediator of immune response(GO:0002440) |
| 0.0 | 0.3 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.8 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 1.8 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.4 | GO:0009749 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 0.2 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.2 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.0 | 0.5 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:1902742 | apoptotic process involved in development(GO:1902742) |
| 0.0 | 0.8 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.7 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.2 | GO:0035545 | determination of left/right asymmetry in diencephalon(GO:0035462) determination of left/right asymmetry in nervous system(GO:0035545) |
| 0.0 | 0.2 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.3 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.1 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:2001286 | optomotor response(GO:0071632) caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 2.2 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.2 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.2 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.3 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.1 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.5 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.1 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.3 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0051645 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.2 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 2.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.4 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.1 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 0.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.3 | GO:0050670 | regulation of mononuclear cell proliferation(GO:0032944) regulation of lymphocyte proliferation(GO:0050670) regulation of leukocyte proliferation(GO:0070663) |
| 0.0 | 28.2 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 0.1 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.0 | 0.5 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) G-protein coupled receptor internalization(GO:0002031) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 0.3 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 2.2 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 8.2 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0051101 | histone H3-K79 methylation(GO:0034729) regulation of DNA binding(GO:0051101) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.0 | 0.1 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.0 | 1.2 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.2 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.2 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.0 | GO:0060039 | pericardium development(GO:0060039) visceral serous pericardium development(GO:0061032) |
| 0.0 | 1.7 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.1 | GO:2000826 | regulation of heart morphogenesis(GO:2000826) |
| 0.0 | 0.3 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.5 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.0 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 1.0 | GO:0050673 | epithelial cell proliferation(GO:0050673) |
| 0.0 | 1.5 | GO:0055123 | digestive system development(GO:0055123) |
| 0.0 | 0.1 | GO:1901071 | glucosamine-containing compound metabolic process(GO:1901071) |
| 0.0 | 0.0 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.2 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.9 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.1 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.1 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.0 | 0.3 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.7 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.3 | GO:0048925 | sensory system development(GO:0048880) lateral line system development(GO:0048925) |
| 0.0 | 2.5 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0008406 | gonad development(GO:0008406) |
| 0.0 | 2.2 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 1.1 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.3 | GO:0031570 | DNA integrity checkpoint(GO:0031570) |
| 0.0 | 0.4 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 | 0.4 | GO:0098542 | defense response to other organism(GO:0098542) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.2 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 0.1 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.1 | GO:0060579 | ventral spinal cord interneuron fate commitment(GO:0060579) |
| 0.0 | 2.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.3 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.2 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.0 | 1.7 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.3 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.2 | GO:0015780 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.1 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.4 | GO:0060563 | neuroepithelial cell differentiation(GO:0060563) |
| 0.0 | 0.0 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.2 | GO:0003094 | glomerular filtration(GO:0003094) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.9 | 2.7 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.9 | 2.6 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.7 | 8.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 1.0 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.3 | 1.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 1.5 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 5.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 1.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.3 | 1.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 1.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 0.7 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.2 | 1.6 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 0.9 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.2 | 1.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.2 | 0.6 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 4.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 1.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 4.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 0.7 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 1.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 0.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 2.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 18.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.7 | GO:0034362 | mature chylomicron(GO:0034359) low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.7 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.8 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 13.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 8.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.9 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 2.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.5 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.4 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 1.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.8 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.1 | 1.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 1.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 2.0 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.4 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.5 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.5 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.8 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.4 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.4 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.1 | 0.4 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 0.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 1.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.8 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.6 | GO:0071256 | translocon complex(GO:0071256) |
| 0.1 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.5 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 1.4 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 3.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 3.3 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 0.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.3 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 7.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 0.3 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 1.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.3 | GO:0030892 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 1.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 1.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 4.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 4.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.4 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 4.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 2.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.6 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 1.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 8.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.3 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 39.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0016234 | inclusion body(GO:0016234) Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.2 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 1.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.8 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.2 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 2.6 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.8 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.2 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.9 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 3.8 | GO:1990904 | ribonucleoprotein complex(GO:1990904) |
| 0.0 | 0.5 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0070822 | Sin3-type complex(GO:0070822) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 1.1 | 5.3 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.6 | 1.9 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.5 | 1.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.5 | 5.2 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.4 | 1.3 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.4 | 2.9 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.4 | 1.6 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.4 | 2.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.4 | 2.0 | GO:0009374 | biotin binding(GO:0009374) |
| 0.4 | 3.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 1.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.4 | 2.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 1.0 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.3 | 1.0 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.3 | 1.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.3 | 1.5 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.3 | 1.8 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 0.9 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.3 | 0.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 3.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 1.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.3 | 0.8 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.3 | 1.3 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.3 | 0.8 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 0.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 0.7 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.2 | 1.9 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 1.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 0.6 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 2.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 1.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.2 | 0.8 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 1.0 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 0.7 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.2 | 1.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 2.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.5 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.2 | 0.4 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.2 | 0.9 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.2 | 1.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 0.5 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 0.7 | GO:0051916 | C-X-C chemokine receptor activity(GO:0016494) granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.2 | 3.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 1.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 3.1 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.2 | 0.5 | GO:0052725 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.2 | 0.5 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.2 | 1.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 0.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 2.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 1.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.2 | 0.9 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 17.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 1.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.9 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 1.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.6 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 0.6 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.1 | 0.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.7 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.6 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.5 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.1 | 30.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 4.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.7 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 1.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.4 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.7 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.1 | 1.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.2 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.1 | 1.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.6 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 0.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.9 | GO:0015157 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 1.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.4 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 0.8 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.6 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.6 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 2.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 1.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.5 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 0.4 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 0.2 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.1 | 0.6 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 0.7 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.3 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 1.6 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 2.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.8 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 1.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 1.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.9 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.7 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.8 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.7 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 1.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 4.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 3.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 11.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 1.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.7 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 1.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.6 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.8 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 3.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.4 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.1 | 0.3 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.6 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 0.3 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.1 | 0.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 2.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.9 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 2.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.9 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.3 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 1.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.9 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 2.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.0 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 1.7 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 0.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 3.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.5 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.3 | GO:0001096 | RNA polymerase II basal transcription factor binding(GO:0001091) TFIIF-class transcription factor binding(GO:0001096) |
| 0.1 | 0.3 | GO:0008026 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 3.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.5 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 3.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 10.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.2 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.1 | 0.3 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.2 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.1 | 0.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.5 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.1 | 0.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 4.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.1 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.2 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 3.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.8 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.2 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 0.5 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 7.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.3 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 4.7 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 4.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 11.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.3 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.3 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 1.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 1.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 2.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.9 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.0 | 0.1 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 1.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 1.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.8 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.0 | 2.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 1.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 4.1 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 0.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 1.1 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0051371 | actinin binding(GO:0042805) muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0051765 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol tetrakisphosphate kinase activity(GO:0051765) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.1 | GO:0010853 | calcium sensitive guanylate cyclase activator activity(GO:0008048) cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 1.1 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 2.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.6 | GO:0016859 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.0 | 0.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 1.4 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.4 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.4 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 34.3 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.8 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.4 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.3 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 2.2 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.1 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 2.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.3 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 0.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.3 | 7.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 1.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 2.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 0.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 3.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.3 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 0.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 0.9 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 2.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 0.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 0.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 0.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 2.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 2.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 1.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 0.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.6 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.5 | 4.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.4 | 2.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.4 | 2.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 1.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 4.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 2.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 1.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 4.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 4.2 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 1.9 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 2.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 11.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.6 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 3.8 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 1.5 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 1.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.5 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 1.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 0.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.6 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 2.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.2 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 4.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.9 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 0.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |