Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
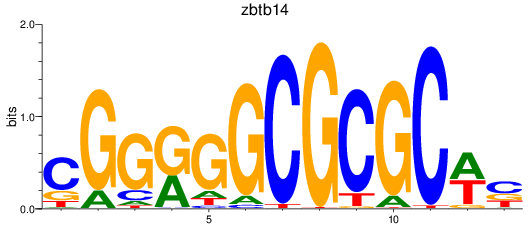
Results for zbtb14
Z-value: 5.04
Transcription factors associated with zbtb14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb14
|
ENSDARG00000098273 | zinc finger and BTB domain containing 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb14 | dr11_v1_chr24_+_41989108_41989108 | 0.83 | 8.5e-02 | Click! |
Activity profile of zbtb14 motif
Sorted Z-values of zbtb14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_25649626 | 7.24 |
ENSDART00000146947
|
tac1
|
tachykinin 1 |
| chr1_-_56223913 | 5.37 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr25_+_14165447 | 5.12 |
ENSDART00000145387
|
shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr1_+_53321878 | 5.02 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr4_-_797831 | 4.86 |
ENSDART00000158970
ENSDART00000170012 |
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
| chr16_-_43026273 | 4.70 |
ENSDART00000156820
ENSDART00000189080 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr23_+_44732863 | 4.35 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr7_+_73332486 | 4.30 |
ENSDART00000174119
ENSDART00000092051 ENSDART00000192388 |
CABZ01081780.1
|
|
| chr21_+_11468934 | 4.26 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr22_-_21150845 | 4.17 |
ENSDART00000027345
|
tmem59l
|
transmembrane protein 59-like |
| chr23_+_45611649 | 4.16 |
ENSDART00000169521
|
dclk2b
|
doublecortin-like kinase 2b |
| chr21_+_11468642 | 4.11 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr1_-_21409877 | 4.02 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr10_+_466926 | 4.00 |
ENSDART00000145220
|
arvcfa
|
ARVCF, delta catenin family member a |
| chr13_+_35746440 | 3.97 |
ENSDART00000187859
|
gpr75
|
G protein-coupled receptor 75 |
| chr2_+_42724404 | 3.83 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr4_+_5741733 | 3.79 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr16_-_43025885 | 3.60 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr4_-_5764255 | 3.58 |
ENSDART00000113864
|
faxca
|
failed axon connections homolog a |
| chr3_-_21280373 | 3.57 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr14_-_2163454 | 3.47 |
ENSDART00000160123
ENSDART00000169653 |
pcdh2ab9
pcdh2ac
|
protocadherin 2 alpha b 9 protocadherin 2 alpha c |
| chr24_+_32176155 | 3.47 |
ENSDART00000003745
|
vim
|
vimentin |
| chr19_-_13733870 | 3.46 |
ENSDART00000177773
|
epb41a
|
erythrocyte membrane protein band 4.1a |
| chr24_+_14713776 | 3.41 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr7_+_39624728 | 3.34 |
ENSDART00000173847
ENSDART00000173845 |
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr11_+_7324704 | 3.28 |
ENSDART00000031937
|
diras1a
|
DIRAS family, GTP-binding RAS-like 1a |
| chr25_+_21324588 | 3.28 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr16_-_26074529 | 3.23 |
ENSDART00000148653
ENSDART00000148923 |
tmem145
|
transmembrane protein 145 |
| chr13_+_27073901 | 3.21 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr18_+_42970208 | 3.14 |
ENSDART00000084454
|
nectin1b
|
nectin cell adhesion molecule 1b |
| chr14_-_9199968 | 3.12 |
ENSDART00000146113
|
arhgef9b
|
Cdc42 guanine nucleotide exchange factor (GEF) 9b |
| chr22_-_372723 | 3.11 |
ENSDART00000112895
|
FNDC10
|
si:zfos-1324h11.5 |
| chr16_+_45456066 | 3.04 |
ENSDART00000093365
|
syngap1b
|
synaptic Ras GTPase activating protein 1b |
| chr24_+_5840748 | 3.02 |
ENSDART00000139191
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr18_-_5692292 | 3.01 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr24_+_5840258 | 3.00 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr7_+_48288762 | 2.99 |
ENSDART00000083569
|
oaz2b
|
ornithine decarboxylase antizyme 2b |
| chr1_-_8428736 | 2.98 |
ENSDART00000138435
ENSDART00000121823 |
syngr3b
|
synaptogyrin 3b |
| chr19_-_5254699 | 2.93 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr13_+_30054996 | 2.92 |
ENSDART00000110061
ENSDART00000186045 |
spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr14_-_34276574 | 2.91 |
ENSDART00000021437
|
gria1a
|
glutamate receptor, ionotropic, AMPA 1a |
| chr5_+_63668735 | 2.91 |
ENSDART00000134261
ENSDART00000097330 |
dnm1b
|
dynamin 1b |
| chr8_+_26868105 | 2.90 |
ENSDART00000005337
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr3_+_17744339 | 2.86 |
ENSDART00000132622
|
znf385c
|
zinc finger protein 385C |
| chr2_+_46032678 | 2.85 |
ENSDART00000184382
ENSDART00000125971 |
gpc1b
|
glypican 1b |
| chr2_+_26179096 | 2.81 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr20_-_40451115 | 2.80 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr19_-_25301711 | 2.79 |
ENSDART00000175739
|
rims3
|
regulating synaptic membrane exocytosis 3 |
| chr13_-_27767330 | 2.77 |
ENSDART00000131631
ENSDART00000112553 ENSDART00000189911 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr20_+_50852356 | 2.76 |
ENSDART00000167517
ENSDART00000168396 |
gphnb
|
gephyrin b |
| chr5_-_22952156 | 2.75 |
ENSDART00000111146
|
si:ch211-26b3.4
|
si:ch211-26b3.4 |
| chr7_-_52334429 | 2.73 |
ENSDART00000187372
|
CR938716.1
|
|
| chr25_-_22639133 | 2.73 |
ENSDART00000073583
|
islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr20_+_474288 | 2.66 |
ENSDART00000026794
|
nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr10_-_13772847 | 2.64 |
ENSDART00000145103
ENSDART00000184491 |
cntfr
|
ciliary neurotrophic factor receptor |
| chr10_-_15405564 | 2.64 |
ENSDART00000020665
|
sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr1_-_20911297 | 2.62 |
ENSDART00000078271
|
cpe
|
carboxypeptidase E |
| chr3_-_36944749 | 2.62 |
ENSDART00000154501
|
cntnap1
|
contactin associated protein 1 |
| chr13_+_30055171 | 2.61 |
ENSDART00000143581
ENSDART00000132027 |
spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr10_-_27223827 | 2.59 |
ENSDART00000185138
|
auts2a
|
autism susceptibility candidate 2a |
| chr2_-_44720551 | 2.58 |
ENSDART00000146380
|
map6d1
|
MAP6 domain containing 1 |
| chr5_-_21030934 | 2.57 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr1_-_38709551 | 2.55 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr3_-_36115339 | 2.55 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr7_+_32722227 | 2.53 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr7_+_74141297 | 2.53 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr5_-_34185497 | 2.53 |
ENSDART00000146321
|
fibcd1
|
fibrinogen C domain containing 1 |
| chr25_-_3979583 | 2.53 |
ENSDART00000124749
|
myrf
|
myelin regulatory factor |
| chr8_+_47897734 | 2.50 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr4_-_133492 | 2.47 |
ENSDART00000171845
|
gpr19
|
G protein-coupled receptor 19 |
| chr17_+_50127648 | 2.43 |
ENSDART00000156460
|
galnt16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr23_-_44226556 | 2.42 |
ENSDART00000149115
|
zgc:158659
|
zgc:158659 |
| chr25_+_4837915 | 2.42 |
ENSDART00000168016
|
gnb5a
|
guanine nucleotide binding protein (G protein), beta 5a |
| chr21_-_25573064 | 2.41 |
ENSDART00000134310
|
CR388166.1
|
|
| chr17_-_45254585 | 2.39 |
ENSDART00000185507
ENSDART00000172080 |
ttbk2a
|
tau tubulin kinase 2a |
| chr19_+_1184878 | 2.38 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr14_-_4682114 | 2.37 |
ENSDART00000014454
|
gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr20_-_33790003 | 2.34 |
ENSDART00000020183
|
fam102bb
|
family with sequence similarity 102, member B, b |
| chr19_-_307246 | 2.33 |
ENSDART00000145916
|
lingo4a
|
leucine rich repeat and Ig domain containing 4a |
| chr2_-_54039293 | 2.33 |
ENSDART00000166013
|
abhd8a
|
abhydrolase domain containing 8a |
| chr13_-_31470439 | 2.32 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr6_-_11780070 | 2.30 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr22_-_12160283 | 2.29 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr8_+_48603398 | 2.23 |
ENSDART00000074900
|
zgc:195023
|
zgc:195023 |
| chr17_-_3986236 | 2.21 |
ENSDART00000188794
ENSDART00000160830 |
PLCB1
|
si:ch1073-140o9.2 |
| chr22_-_29586608 | 2.18 |
ENSDART00000059869
|
adra2a
|
adrenoceptor alpha 2A |
| chr6_-_13308813 | 2.17 |
ENSDART00000065372
|
kcnj3b
|
potassium inwardly-rectifying channel, subfamily J, member 3b |
| chr21_+_39423974 | 2.16 |
ENSDART00000031470
|
pafah1b1b
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b |
| chr23_+_45611980 | 2.14 |
ENSDART00000181582
|
dclk2b
|
doublecortin-like kinase 2b |
| chr22_+_4488454 | 2.14 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr12_+_21299338 | 2.07 |
ENSDART00000074540
ENSDART00000133188 |
ca10a
|
carbonic anhydrase Xa |
| chr12_+_12112384 | 2.06 |
ENSDART00000152431
|
grid1b
|
glutamate receptor, ionotropic, delta 1b |
| chr24_-_25673405 | 2.05 |
ENSDART00000186081
ENSDART00000110241 ENSDART00000142351 |
cnksr2a
|
connector enhancer of kinase suppressor of Ras 2a |
| chr2_-_24462277 | 2.05 |
ENSDART00000033922
|
kcnn1a
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1a |
| chr17_-_24717925 | 2.03 |
ENSDART00000153725
|
si:ch211-15d5.11
|
si:ch211-15d5.11 |
| chr7_+_14632157 | 2.03 |
ENSDART00000161264
|
ntrk3b
|
neurotrophic tyrosine kinase, receptor, type 3b |
| chr3_+_32789605 | 2.03 |
ENSDART00000171895
|
tbc1d10b
|
TBC1 domain family, member 10b |
| chr14_+_11103718 | 2.01 |
ENSDART00000161311
|
nexmifb
|
neurite extension and migration factor b |
| chr7_-_49594995 | 2.00 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr9_+_53707240 | 1.99 |
ENSDART00000171490
|
PCDH8
|
si:ch211-199f5.1 |
| chr3_-_1204341 | 1.98 |
ENSDART00000089646
|
fam234b
|
family with sequence similarity 234, member B |
| chr4_+_2482046 | 1.98 |
ENSDART00000103371
|
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr5_-_31712399 | 1.96 |
ENSDART00000141328
|
pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr17_-_20287530 | 1.94 |
ENSDART00000078703
ENSDART00000191289 |
add3b
|
adducin 3 (gamma) b |
| chr12_+_26621906 | 1.92 |
ENSDART00000158440
ENSDART00000046959 |
arhgap12b
|
Rho GTPase activating protein 12b |
| chr11_+_15931188 | 1.91 |
ENSDART00000165768
ENSDART00000081098 |
draxin
|
dorsal inhibitory axon guidance protein |
| chr11_-_37589293 | 1.86 |
ENSDART00000172989
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr22_+_3045495 | 1.85 |
ENSDART00000164061
|
LO017843.1
|
|
| chr10_-_25069155 | 1.84 |
ENSDART00000078226
ENSDART00000181941 |
mtnr1bb
|
melatonin receptor 1Bb |
| chr22_+_38978084 | 1.83 |
ENSDART00000025482
|
arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr1_-_22834824 | 1.82 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr7_-_47850702 | 1.82 |
ENSDART00000109511
|
si:ch211-186j3.6
|
si:ch211-186j3.6 |
| chr25_-_3979288 | 1.82 |
ENSDART00000157117
|
myrf
|
myelin regulatory factor |
| chr24_+_39227519 | 1.81 |
ENSDART00000184611
ENSDART00000193494 ENSDART00000190728 ENSDART00000168705 |
MPRIP
|
si:ch73-103b11.2 |
| chr1_+_20635190 | 1.78 |
ENSDART00000145418
ENSDART00000148518 ENSDART00000139461 ENSDART00000102969 ENSDART00000166479 |
spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr18_-_44527124 | 1.78 |
ENSDART00000189471
|
aplp2
|
amyloid beta (A4) precursor-like protein 2 |
| chr10_-_21054059 | 1.78 |
ENSDART00000139733
|
pcdh1a
|
protocadherin 1a |
| chr1_-_50859053 | 1.77 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr14_-_2400668 | 1.76 |
ENSDART00000172717
ENSDART00000182882 |
si:ch73-233f7.1
|
si:ch73-233f7.1 |
| chr18_-_44526940 | 1.75 |
ENSDART00000077125
|
aplp2
|
amyloid beta (A4) precursor-like protein 2 |
| chr12_-_30032188 | 1.74 |
ENSDART00000042514
|
atrnl1b
|
attractin-like 1b |
| chr24_+_16393302 | 1.74 |
ENSDART00000188670
ENSDART00000081759 ENSDART00000177790 |
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr13_+_29773153 | 1.74 |
ENSDART00000144443
ENSDART00000133796 ENSDART00000141310 ENSDART00000139782 |
pax2a
|
paired box 2a |
| chr24_+_4977862 | 1.74 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr17_+_51764310 | 1.74 |
ENSDART00000157171
|
si:ch211-168d23.3
|
si:ch211-168d23.3 |
| chr22_-_22416337 | 1.72 |
ENSDART00000142947
ENSDART00000089569 |
camsap2a
|
calmodulin regulated spectrin-associated protein family, member 2a |
| chr19_+_4463389 | 1.72 |
ENSDART00000168805
|
kcnk9
|
potassium channel, subfamily K, member 9 |
| chr19_+_14059349 | 1.71 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr24_-_39354829 | 1.70 |
ENSDART00000169108
|
kcnj12a
|
potassium inwardly-rectifying channel, subfamily J, member 12a |
| chr18_+_16125852 | 1.69 |
ENSDART00000061106
|
bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr14_+_8947282 | 1.67 |
ENSDART00000047993
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr16_+_32559821 | 1.67 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr14_-_25577094 | 1.65 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr11_+_29856032 | 1.65 |
ENSDART00000079150
|
grpr
|
gastrin-releasing peptide receptor |
| chr2_+_40294313 | 1.64 |
ENSDART00000037292
|
epha4b
|
eph receptor A4b |
| chr1_-_23157583 | 1.63 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr16_+_52771199 | 1.63 |
ENSDART00000111383
|
baalca
|
brain and acute leukemia, cytoplasmic a |
| chr13_-_29421331 | 1.62 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr24_-_35699595 | 1.62 |
ENSDART00000167990
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr12_+_1398404 | 1.62 |
ENSDART00000026303
|
rasd1
|
RAS, dexamethasone-induced 1 |
| chr2_-_44398611 | 1.59 |
ENSDART00000146192
|
pip4p1a
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1a |
| chr3_-_21348478 | 1.59 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr2_-_9259283 | 1.59 |
ENSDART00000133092
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr24_-_6501211 | 1.59 |
ENSDART00000186241
ENSDART00000109040 ENSDART00000136154 |
gpr158a
|
G protein-coupled receptor 158a |
| chr2_-_30324610 | 1.58 |
ENSDART00000185422
|
jph1b
|
junctophilin 1b |
| chr4_-_9810999 | 1.58 |
ENSDART00000146858
|
si:dkeyp-27e10.3
|
si:dkeyp-27e10.3 |
| chr1_-_47771742 | 1.57 |
ENSDART00000101079
|
neurl1aa
|
neuralized E3 ubiquitin protein ligase 1Aa |
| chr5_+_28973264 | 1.55 |
ENSDART00000005638
|
stxbp1b
|
syntaxin binding protein 1b |
| chr12_+_47698356 | 1.55 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr1_-_57280585 | 1.54 |
ENSDART00000152220
|
si:dkey-27j5.5
|
si:dkey-27j5.5 |
| chr2_+_38742338 | 1.54 |
ENSDART00000177290
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr9_-_14504834 | 1.53 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr14_-_48588422 | 1.52 |
ENSDART00000161147
|
si:ch211-154c21.1
|
si:ch211-154c21.1 |
| chr10_+_19310688 | 1.50 |
ENSDART00000141364
|
lrrtm4l1
|
leucine rich repeat transmembrane neuronal 4 like 1 |
| chr25_-_20381107 | 1.50 |
ENSDART00000067454
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr8_+_23916647 | 1.50 |
ENSDART00000143152
|
cpne5a
|
copine Va |
| chr20_+_18209895 | 1.50 |
ENSDART00000111063
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr10_-_25328814 | 1.49 |
ENSDART00000123820
|
tmem135
|
transmembrane protein 135 |
| chr9_+_21843915 | 1.48 |
ENSDART00000101977
|
kctd12.1
|
potassium channel tetramerisation domain containing 12.1 |
| chr25_-_20381271 | 1.48 |
ENSDART00000142665
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr20_+_38322444 | 1.47 |
ENSDART00000161741
ENSDART00000132241 ENSDART00000148936 ENSDART00000149623 |
stum
|
stum, mechanosensory transduction mediator homolog |
| chr4_-_2196798 | 1.47 |
ENSDART00000110178
ENSDART00000149330 |
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr5_+_28972935 | 1.46 |
ENSDART00000193274
|
stxbp1b
|
syntaxin binding protein 1b |
| chr4_-_3145359 | 1.46 |
ENSDART00000112210
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr12_+_21298317 | 1.44 |
ENSDART00000178562
|
ca10a
|
carbonic anhydrase Xa |
| chr24_+_41931585 | 1.42 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr18_-_6856380 | 1.41 |
ENSDART00000175747
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr9_+_8929764 | 1.40 |
ENSDART00000102562
|
ankrd10b
|
ankyrin repeat domain 10b |
| chr14_-_9056066 | 1.35 |
ENSDART00000139669
ENSDART00000138758 ENSDART00000041099 |
sybl1
|
synaptobrevin-like 1 |
| chr14_-_7306983 | 1.35 |
ENSDART00000158914
|
si:ch211-51f19.1
|
si:ch211-51f19.1 |
| chr5_+_64277604 | 1.35 |
ENSDART00000111282
|
qsox2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr22_-_29922872 | 1.35 |
ENSDART00000020249
|
dusp5
|
dual specificity phosphatase 5 |
| chr10_+_23060391 | 1.35 |
ENSDART00000079711
|
slc25a1a
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1a |
| chr12_-_7607114 | 1.34 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr20_-_35246150 | 1.33 |
ENSDART00000090549
|
fzd3a
|
frizzled class receptor 3a |
| chr24_-_35699444 | 1.33 |
ENSDART00000166567
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr17_-_2535682 | 1.30 |
ENSDART00000155227
|
ccdc9b
|
coiled-coil domain containing 9B |
| chr10_-_37075361 | 1.30 |
ENSDART00000132023
|
myo18aa
|
myosin XVIIIAa |
| chr6_-_957830 | 1.30 |
ENSDART00000090019
ENSDART00000184286 |
zeb2b
|
zinc finger E-box binding homeobox 2b |
| chr7_+_73593814 | 1.29 |
ENSDART00000110544
|
znf219
|
zinc finger protein 219 |
| chr18_-_6855991 | 1.29 |
ENSDART00000135206
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr18_-_46882056 | 1.28 |
ENSDART00000108574
|
gramd1bb
|
GRAM domain containing 1Bb |
| chr23_-_10914275 | 1.28 |
ENSDART00000112965
|
pdzrn3a
|
PDZ domain containing RING finger 3a |
| chr4_-_12007404 | 1.28 |
ENSDART00000092250
|
btbd11a
|
BTB (POZ) domain containing 11a |
| chr17_+_15983247 | 1.28 |
ENSDART00000154275
|
clmn
|
calmin |
| chr1_+_9004719 | 1.28 |
ENSDART00000006211
ENSDART00000137211 |
prkcba
|
protein kinase C, beta a |
| chr24_-_27879182 | 1.27 |
ENSDART00000105778
|
hs6st1b
|
heparan sulfate 6-O-sulfotransferase 1b |
| chr15_+_36941490 | 1.27 |
ENSDART00000172664
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr10_+_45248326 | 1.25 |
ENSDART00000159797
|
zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr25_-_12203952 | 1.24 |
ENSDART00000158204
ENSDART00000091727 |
ntrk3a
|
neurotrophic tyrosine kinase, receptor, type 3a |
| chr24_-_38816725 | 1.24 |
ENSDART00000063231
|
nog2
|
noggin 2 |
| chr22_+_28803739 | 1.24 |
ENSDART00000129476
ENSDART00000189726 |
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr7_-_19369002 | 1.24 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr24_+_4978055 | 1.23 |
ENSDART00000045813
|
zic4
|
zic family member 4 |
| chr6_+_4539953 | 1.22 |
ENSDART00000025031
|
pou4f1
|
POU class 4 homeobox 1 |
| chr14_-_9355177 | 1.20 |
ENSDART00000138535
|
fam46d
|
family with sequence similarity 46, member D |
| chr25_+_3677650 | 1.20 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr24_+_35881124 | 1.20 |
ENSDART00000143015
|
klhl14
|
kelch-like family member 14 |
| chr7_-_24236364 | 1.19 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr19_+_41075488 | 1.18 |
ENSDART00000138687
|
ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb14
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 1.0 | 3.1 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.9 | 2.8 | GO:0072579 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.9 | 2.6 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.9 | 7.8 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.8 | 4.2 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.6 | 2.9 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.5 | 1.6 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.5 | 2.0 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.5 | 4.3 | GO:0043217 | myelin maintenance(GO:0043217) |
| 0.5 | 2.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.5 | 8.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.4 | 1.8 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.4 | 2.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.4 | 1.7 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.4 | 3.0 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.4 | 2.6 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.4 | 5.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.4 | 1.1 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) regulation of apoptotic cell clearance(GO:2000425) |
| 0.3 | 1.3 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.3 | 1.0 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.3 | 1.7 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.3 | 2.0 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.3 | 1.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 11.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.3 | 0.9 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.3 | 1.4 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.3 | 2.6 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.3 | 1.5 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.3 | 5.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.2 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.2 | 4.4 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 1.7 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.2 | 2.8 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.2 | 1.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.2 | 1.0 | GO:0060544 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.2 | 1.0 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.2 | 1.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 9.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 1.9 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.2 | 3.3 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.2 | 2.2 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.2 | 0.8 | GO:0045056 | transcytosis(GO:0045056) |
| 0.2 | 0.9 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 1.7 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.2 | 1.7 | GO:0030431 | sleep(GO:0030431) |
| 0.2 | 2.9 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.7 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 1.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.7 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 0.7 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 1.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.5 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 2.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.8 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.9 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 3.0 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.1 | 1.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.3 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 4.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 4.7 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.4 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 2.7 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 2.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 2.4 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 1.1 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 2.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 3.4 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 0.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.4 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 0.3 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 2.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 1.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.1 | 0.4 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.1 | 1.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.2 | GO:0009193 | ribonucleoside diphosphate catabolic process(GO:0009191) pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 2.5 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.7 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 2.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 3.9 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 1.3 | GO:0072176 | pronephric duct development(GO:0039022) nephric duct development(GO:0072176) |
| 0.1 | 1.3 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 4.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 1.4 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 2.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 7.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 3.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.3 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 2.7 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 1.5 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 4.6 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 1.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 2.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.2 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.2 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.4 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.9 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.4 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.4 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 5.9 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 1.3 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 2.3 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 2.0 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 2.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.3 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.7 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 1.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.5 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 1.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 1.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 1.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.3 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.4 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 5.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 1.1 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 1.1 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 5.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 3.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.0 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 1.3 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.2 | GO:1901571 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.0 | 1.0 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 1.0 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 2.0 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.1 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 1.6 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 1.9 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0072380 | TRC complex(GO:0072380) |
| 0.5 | 7.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.5 | 2.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.4 | 4.4 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.4 | 5.9 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.4 | 1.8 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.3 | 2.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.3 | 7.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 7.4 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.2 | 4.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 2.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 6.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 2.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.7 | GO:1990752 | microtubule end(GO:1990752) |
| 0.2 | 0.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 6.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 2.9 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 2.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 3.8 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.1 | 0.5 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 2.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.5 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 3.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.1 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.1 | 4.1 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 0.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 8.7 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.1 | 2.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 8.9 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 1.7 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.0 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 7.6 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 4.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.6 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 8.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.4 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 2.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 6.3 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.3 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 18.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.6 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 5.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 8.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.4 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.7 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.7 | GO:0036064 | ciliary basal body(GO:0036064) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 1.0 | 2.9 | GO:0072591 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.9 | 2.8 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.7 | 3.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.5 | 3.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 2.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.5 | 6.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.4 | 1.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.4 | 2.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 7.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.4 | 2.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.4 | 3.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 1.8 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 7.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.3 | 0.9 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.3 | 1.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 2.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 8.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 2.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 2.4 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.2 | 4.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 1.3 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 1.6 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 1.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 0.8 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.2 | 1.0 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 1.0 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 1.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 1.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 0.9 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 2.4 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 0.7 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 1.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 1.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 2.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.2 | 1.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.4 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.1 | 2.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 2.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 3.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.9 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 2.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.6 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 2.1 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.1 | 1.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 2.8 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 2.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 5.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.9 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.5 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 3.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.5 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 1.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 4.0 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 1.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 1.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.9 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 4.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 1.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 1.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 3.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 2.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 6.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 1.3 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 1.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 4.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.7 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 1.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 3.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 2.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 11.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 3.6 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 1.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 3.5 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 7.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.5 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.8 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 4.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 2.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 9.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 3.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 1.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 1.0 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 2.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 2.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 2.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 2.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 11.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.8 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 1.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.8 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 3.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.5 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 1.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 2.1 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.8 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 1.1 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 0.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |