Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
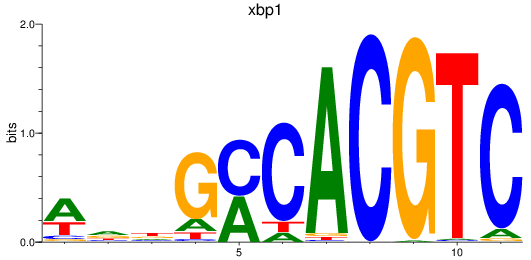
Results for xbp1
Z-value: 1.11
Transcription factors associated with xbp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
xbp1
|
ENSDARG00000035622 | X-box binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| xbp1 | dr11_v1_chr5_-_15948833_15948920 | 0.83 | 8.3e-02 | Click! |
Activity profile of xbp1 motif
Sorted Z-values of xbp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23984755 | 0.65 |
ENSDART00000145328
|
apoc2
|
apolipoprotein C-II |
| chr4_+_68562464 | 0.60 |
ENSDART00000192954
|
BX548011.4
|
|
| chr16_+_27224369 | 0.60 |
ENSDART00000136980
|
sec61b
|
Sec61 translocon beta subunit |
| chr16_+_23984179 | 0.44 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr7_+_10610791 | 0.43 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr3_-_25377163 | 0.42 |
ENSDART00000055490
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr16_-_46660680 | 0.42 |
ENSDART00000159209
ENSDART00000191929 |
tmem176l.4
|
transmembrane protein 176l.4 |
| chr12_-_990149 | 0.37 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr21_+_10701834 | 0.33 |
ENSDART00000192473
|
lman1
|
lectin, mannose-binding, 1 |
| chr6_+_13506841 | 0.32 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr8_+_11471350 | 0.32 |
ENSDART00000092355
ENSDART00000136184 |
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr21_+_10702031 | 0.31 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr22_-_817479 | 0.31 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr24_+_14214831 | 0.30 |
ENSDART00000004664
|
tram1
|
translocation associated membrane protein 1 |
| chr7_-_19614916 | 0.30 |
ENSDART00000169029
|
zgc:194655
|
zgc:194655 |
| chr16_-_26676685 | 0.30 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr22_+_38194151 | 0.29 |
ENSDART00000121965
|
cp
|
ceruloplasmin |
| chr4_-_13613148 | 0.29 |
ENSDART00000067164
ENSDART00000111247 |
irf5
|
interferon regulatory factor 5 |
| chr1_-_23308225 | 0.28 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr25_+_5983430 | 0.28 |
ENSDART00000074814
|
ppib
|
peptidylprolyl isomerase B (cyclophilin B) |
| chr19_-_30404096 | 0.28 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr8_-_14554785 | 0.28 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr19_-_11238620 | 0.27 |
ENSDART00000148697
ENSDART00000149169 |
ssr2
|
signal sequence receptor, beta |
| chr19_-_30403922 | 0.27 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr23_-_18572521 | 0.27 |
ENSDART00000138599
|
ssr4
|
signal sequence receptor, delta |
| chr8_+_5024468 | 0.25 |
ENSDART00000030938
|
adra1aa
|
adrenoceptor alpha 1Aa |
| chr16_+_35918463 | 0.24 |
ENSDART00000160608
|
sh3d21
|
SH3 domain containing 21 |
| chr23_-_18572685 | 0.24 |
ENSDART00000047175
|
ssr4
|
signal sequence receptor, delta |
| chr6_+_6779114 | 0.24 |
ENSDART00000163493
ENSDART00000143359 |
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr7_+_19615056 | 0.24 |
ENSDART00000124752
ENSDART00000190297 |
si:ch211-212k18.15
|
si:ch211-212k18.15 |
| chr24_-_42090635 | 0.24 |
ENSDART00000166413
|
ssr1
|
signal sequence receptor, alpha |
| chr18_+_7553950 | 0.24 |
ENSDART00000193420
ENSDART00000062143 |
zgc:77650
|
zgc:77650 |
| chr17_+_32360673 | 0.24 |
ENSDART00000155519
|
si:ch211-139d20.3
|
si:ch211-139d20.3 |
| chr5_+_58550291 | 0.23 |
ENSDART00000184983
ENSDART00000044803 |
pou2f3
|
POU class 2 homeobox 3 |
| chr10_-_34915886 | 0.23 |
ENSDART00000141201
ENSDART00000002166 |
ccna1
|
cyclin A1 |
| chr25_+_3759553 | 0.23 |
ENSDART00000180601
ENSDART00000055845 ENSDART00000157050 ENSDART00000153905 |
thoc5
|
THO complex 5 |
| chr10_-_22150419 | 0.22 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr19_+_18903533 | 0.22 |
ENSDART00000157523
ENSDART00000166562 |
slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr17_+_53156530 | 0.22 |
ENSDART00000126277
ENSDART00000156774 |
dph6
|
diphthamine biosynthesis 6 |
| chr1_-_45049603 | 0.22 |
ENSDART00000023336
|
rps6
|
ribosomal protein S6 |
| chr24_-_33291784 | 0.22 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr5_+_69868911 | 0.21 |
ENSDART00000014649
ENSDART00000188215 ENSDART00000167385 |
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr11_+_44503774 | 0.21 |
ENSDART00000169295
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr13_-_47403154 | 0.21 |
ENSDART00000114318
|
bcl2l11
|
BCL2 like 11 |
| chr11_-_7261717 | 0.21 |
ENSDART00000128959
|
zgc:113223
|
zgc:113223 |
| chr5_+_65087226 | 0.20 |
ENSDART00000183187
|
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr7_+_44715224 | 0.20 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr21_-_22693730 | 0.20 |
ENSDART00000158342
|
gig2d
|
grass carp reovirus (GCRV)-induced gene 2d |
| chr6_-_40071899 | 0.20 |
ENSDART00000034730
|
sec61a1
|
Sec61 translocon alpha 1 subunit |
| chr25_-_37262220 | 0.20 |
ENSDART00000153789
ENSDART00000155182 |
rfwd3
|
ring finger and WD repeat domain 3 |
| chr17_+_24446705 | 0.19 |
ENSDART00000163221
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr10_-_44305180 | 0.19 |
ENSDART00000187552
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr23_+_43638982 | 0.18 |
ENSDART00000168646
|
slc10a7
|
solute carrier family 10, member 7 |
| chr22_-_10152629 | 0.18 |
ENSDART00000144209
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr5_+_65086856 | 0.18 |
ENSDART00000169209
ENSDART00000162409 |
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr2_-_42492201 | 0.17 |
ENSDART00000180762
ENSDART00000009093 |
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr4_+_77957611 | 0.17 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr5_+_2002804 | 0.17 |
ENSDART00000064088
|
vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr18_-_34170918 | 0.17 |
ENSDART00000015079
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr22_+_31059919 | 0.17 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr5_+_65086668 | 0.16 |
ENSDART00000183746
|
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr4_-_19693978 | 0.16 |
ENSDART00000100974
ENSDART00000040405 |
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr1_-_9486214 | 0.16 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr2_+_36608387 | 0.16 |
ENSDART00000159541
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr2_-_55298075 | 0.16 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr12_-_16452200 | 0.16 |
ENSDART00000037601
|
rpp30
|
ribonuclease P/MRP 30 subunit |
| chr20_-_26929685 | 0.15 |
ENSDART00000132556
|
ftr79
|
finTRIM family, member 79 |
| chr19_+_43119698 | 0.15 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr14_-_45464112 | 0.15 |
ENSDART00000173209
|
ehd1a
|
EH-domain containing 1a |
| chr10_+_16165533 | 0.15 |
ENSDART00000065045
|
prrc1
|
proline-rich coiled-coil 1 |
| chr4_-_17725008 | 0.15 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr24_-_26485098 | 0.15 |
ENSDART00000135496
ENSDART00000009609 ENSDART00000133782 ENSDART00000141029 ENSDART00000113739 |
eif5a
|
eukaryotic translation initiation factor 5A |
| chr22_-_17781213 | 0.14 |
ENSDART00000137984
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr3_+_28860283 | 0.14 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr10_+_39212898 | 0.14 |
ENSDART00000159501
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr2_-_9527129 | 0.14 |
ENSDART00000157422
ENSDART00000004398 |
cope
|
coatomer protein complex, subunit epsilon |
| chr6_-_54816567 | 0.14 |
ENSDART00000150079
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr18_-_34171280 | 0.14 |
ENSDART00000122321
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr9_+_25840720 | 0.14 |
ENSDART00000024572
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr20_-_30377221 | 0.14 |
ENSDART00000126229
|
rps7
|
ribosomal protein S7 |
| chr6_-_42388608 | 0.14 |
ENSDART00000049425
|
sec61a1l
|
Sec61 translocon alpha 1 subunit, like |
| chr19_+_1414604 | 0.14 |
ENSDART00000159024
|
btr29
|
bloodthirsty-related gene family, member 29 |
| chr4_-_5912951 | 0.14 |
ENSDART00000169439
|
arl1
|
ADP-ribosylation factor-like 1 |
| chr16_-_5115993 | 0.14 |
ENSDART00000138654
|
ttk
|
ttk protein kinase |
| chr1_+_52754085 | 0.14 |
ENSDART00000185420
|
wu:fc75a09
|
wu:fc75a09 |
| chr11_+_45286911 | 0.14 |
ENSDART00000181763
|
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr11_-_2700397 | 0.14 |
ENSDART00000082511
|
si:ch211-160o17.6
|
si:ch211-160o17.6 |
| chr12_+_30788912 | 0.14 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr12_+_30789611 | 0.13 |
ENSDART00000181501
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr9_-_2594410 | 0.13 |
ENSDART00000188306
ENSDART00000164276 |
sp9
|
sp9 transcription factor |
| chr4_+_25651720 | 0.13 |
ENSDART00000100693
ENSDART00000100717 |
acot16
|
acyl-CoA thioesterase 16 |
| chr23_-_24493768 | 0.13 |
ENSDART00000135559
ENSDART00000046933 |
sult1st5
|
sulfotransferase family 1, cytosolic sulfotransferase 5 |
| chr11_+_6431133 | 0.13 |
ENSDART00000190742
|
FO904935.1
|
|
| chr11_-_6420917 | 0.13 |
ENSDART00000193717
|
FO681393.2
|
|
| chr19_+_14113886 | 0.12 |
ENSDART00000169343
|
kdf1b
|
keratinocyte differentiation factor 1b |
| chr25_-_774350 | 0.12 |
ENSDART00000166321
ENSDART00000160386 |
IRAK4
|
interleukin 1 receptor associated kinase 4 |
| chr6_-_35052145 | 0.12 |
ENSDART00000073970
ENSDART00000185790 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr1_+_38142354 | 0.12 |
ENSDART00000179352
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr9_-_27748868 | 0.12 |
ENSDART00000190306
|
tbccd1
|
TBCC domain containing 1 |
| chr20_-_40758410 | 0.12 |
ENSDART00000183031
|
cx34.5
|
connexin 34.5 |
| chr2_-_7757273 | 0.12 |
ENSDART00000136074
|
b3gnt5b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5b |
| chr12_-_4206869 | 0.12 |
ENSDART00000106572
|
si:dkey-32n7.9
|
si:dkey-32n7.9 |
| chr4_-_77979432 | 0.11 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr17_+_24446353 | 0.11 |
ENSDART00000140467
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr5_+_24543862 | 0.11 |
ENSDART00000029699
|
atp6v0a2b
|
ATPase H+ transporting V0 subunit a2b |
| chr1_-_9485939 | 0.11 |
ENSDART00000157814
|
micall2b
|
mical-like 2b |
| chr24_-_982443 | 0.11 |
ENSDART00000063151
|
napga
|
N-ethylmaleimide-sensitive factor attachment protein, gamma a |
| chr2_-_22363460 | 0.11 |
ENSDART00000158486
|
selenof
|
selenoprotein F |
| chr5_+_21211135 | 0.11 |
ENSDART00000088492
|
bmp10
|
bone morphogenetic protein 10 |
| chr11_+_45287541 | 0.11 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr5_-_41841675 | 0.11 |
ENSDART00000141683
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr12_+_31783066 | 0.11 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr9_+_54237100 | 0.10 |
ENSDART00000148928
|
rbm27
|
RNA binding motif protein 27 |
| chr17_+_24446533 | 0.10 |
ENSDART00000131417
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr14_-_16082806 | 0.10 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr6_-_52348562 | 0.10 |
ENSDART00000142565
ENSDART00000145369 ENSDART00000016890 |
eif6
|
eukaryotic translation initiation factor 6 |
| chr22_+_3238474 | 0.10 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr4_+_13568469 | 0.10 |
ENSDART00000171235
ENSDART00000136152 |
calua
|
calumenin a |
| chr7_-_56606752 | 0.10 |
ENSDART00000138714
|
sult5a1
|
sulfotransferase family 5A, member 1 |
| chr24_-_11069237 | 0.10 |
ENSDART00000127398
|
CR753886.1
|
|
| chr25_-_3034668 | 0.10 |
ENSDART00000053405
|
scamp2
|
secretory carrier membrane protein 2 |
| chr18_-_18587745 | 0.09 |
ENSDART00000191973
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr10_+_39212601 | 0.09 |
ENSDART00000168778
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr5_+_20453874 | 0.09 |
ENSDART00000124545
ENSDART00000008402 |
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr14_+_31566517 | 0.09 |
ENSDART00000002684
|
ints6l
|
integrator complex subunit 6 like |
| chr1_+_38142715 | 0.09 |
ENSDART00000079928
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr1_-_53277413 | 0.09 |
ENSDART00000150579
|
znf330
|
zinc finger protein 330 |
| chr19_-_29832876 | 0.09 |
ENSDART00000005119
|
eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr19_-_5103141 | 0.09 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr6_+_3864040 | 0.09 |
ENSDART00000013743
|
gorasp2
|
golgi reassembly stacking protein 2 |
| chr21_-_8054977 | 0.08 |
ENSDART00000130508
|
si:dkey-163m14.4
|
si:dkey-163m14.4 |
| chr19_+_26640096 | 0.08 |
ENSDART00000067793
|
ints3
|
integrator complex subunit 3 |
| chr9_+_20526472 | 0.08 |
ENSDART00000166457
ENSDART00000141775 |
trim45
|
tripartite motif containing 45 |
| chr6_-_35052388 | 0.08 |
ENSDART00000181000
ENSDART00000170116 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr16_+_6727195 | 0.08 |
ENSDART00000148625
|
znf407
|
zinc finger protein 407 |
| chr6_-_54815886 | 0.08 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr19_-_5103313 | 0.08 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr12_-_2869565 | 0.08 |
ENSDART00000152772
|
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr4_-_5913338 | 0.08 |
ENSDART00000183590
|
arl1
|
ADP-ribosylation factor-like 1 |
| chr22_+_2844865 | 0.08 |
ENSDART00000139123
|
si:dkey-20i20.4
|
si:dkey-20i20.4 |
| chr3_-_55092051 | 0.08 |
ENSDART00000053077
|
hbba2
|
hemoglobin, beta adult 2 |
| chr9_+_38273233 | 0.08 |
ENSDART00000142853
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr10_-_3427589 | 0.08 |
ENSDART00000133452
ENSDART00000037183 |
tmed2
|
transmembrane p24 trafficking protein 2 |
| chr8_-_44463985 | 0.07 |
ENSDART00000016845
|
mhc1lba
|
major histocompatibility complex class I LBA |
| chr3_-_36209936 | 0.07 |
ENSDART00000175208
|
csnk1da
|
casein kinase 1, delta a |
| chr9_+_21165017 | 0.07 |
ENSDART00000145933
ENSDART00000142985 |
si:rp71-68n21.9
|
si:rp71-68n21.9 |
| chr9_+_23825440 | 0.07 |
ENSDART00000138470
|
ints6
|
integrator complex subunit 6 |
| chr8_+_47219107 | 0.07 |
ENSDART00000146018
ENSDART00000075068 |
mthfr
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr1_-_156375 | 0.07 |
ENSDART00000160221
|
pcid2
|
PCI domain containing 2 |
| chr25_-_34512102 | 0.07 |
ENSDART00000087450
|
klf13
|
Kruppel-like factor 13 |
| chr15_-_29326254 | 0.07 |
ENSDART00000114492
|
si:dkey-52l18.4
|
si:dkey-52l18.4 |
| chr18_+_2593756 | 0.07 |
ENSDART00000158022
|
p2ry2.3
|
purinergic receptor P2Y2, tandem duplicate 3 |
| chr1_-_26292897 | 0.07 |
ENSDART00000112899
ENSDART00000185410 |
cxxc4
|
CXXC finger 4 |
| chr22_-_11078988 | 0.06 |
ENSDART00000126664
ENSDART00000006927 |
use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr5_-_56964547 | 0.06 |
ENSDART00000074400
|
tia1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr5_+_69278089 | 0.06 |
ENSDART00000136656
|
selenom
|
selenoprotein M |
| chr1_+_7956030 | 0.06 |
ENSDART00000159655
|
CR855320.1
|
|
| chr3_+_29510818 | 0.06 |
ENSDART00000055407
ENSDART00000193743 ENSDART00000123619 |
rac2
|
Rac family small GTPase 2 |
| chr17_-_6765877 | 0.06 |
ENSDART00000193102
|
CR352340.1
|
|
| chr16_-_42750295 | 0.06 |
ENSDART00000176570
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr17_+_24445818 | 0.06 |
ENSDART00000139963
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr10_-_5857548 | 0.06 |
ENSDART00000166933
|
si:ch211-281k19.2
|
si:ch211-281k19.2 |
| chr17_+_1514711 | 0.06 |
ENSDART00000165641
|
akt1
|
v-akt murine thymoma viral oncogene homolog 1 |
| chr1_+_44852497 | 0.06 |
ENSDART00000135067
|
kdm2aa
|
lysine (K)-specific demethylase 2Aa |
| chr18_-_7481036 | 0.06 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr21_+_28724099 | 0.06 |
ENSDART00000138017
|
puraa
|
purine-rich element binding protein Aa |
| chr5_+_4366431 | 0.05 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr5_-_30516646 | 0.05 |
ENSDART00000014666
|
arcn1a
|
archain 1a |
| chr2_+_37176814 | 0.05 |
ENSDART00000048277
ENSDART00000176827 |
copa
|
coatomer protein complex, subunit alpha |
| chr5_-_24219344 | 0.05 |
ENSDART00000011878
|
eif4a1b
|
eukaryotic translation initiation factor 4A1B |
| chr13_-_44808783 | 0.05 |
ENSDART00000099984
|
glo1
|
glyoxalase 1 |
| chr12_+_1592146 | 0.05 |
ENSDART00000184575
ENSDART00000192902 |
SLC39A11
|
solute carrier family 39 member 11 |
| chr20_+_29565906 | 0.05 |
ENSDART00000062383
|
ywhaqa
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide a |
| chr3_-_34586403 | 0.05 |
ENSDART00000151515
|
sept9a
|
septin 9a |
| chr8_-_31701157 | 0.05 |
ENSDART00000141799
|
fbxo4
|
F-box protein 4 |
| chr3_+_3610140 | 0.05 |
ENSDART00000104977
|
pick1
|
protein interacting with prkca 1 |
| chr23_+_35672542 | 0.05 |
ENSDART00000046268
|
pmelb
|
premelanosome protein b |
| chr19_-_46219035 | 0.05 |
ENSDART00000170601
|
si:ch1073-228h2.2
|
si:ch1073-228h2.2 |
| chr13_-_38088627 | 0.05 |
ENSDART00000175268
|
CT027676.1
|
|
| chr6_+_23026714 | 0.05 |
ENSDART00000124948
|
srp68
|
signal recognition particle 68 |
| chr18_+_6706140 | 0.05 |
ENSDART00000111343
|
lmf2a
|
lipase maturation factor 2a |
| chr22_+_1517318 | 0.05 |
ENSDART00000160406
|
si:ch211-255f4.5
|
si:ch211-255f4.5 |
| chr12_-_35582521 | 0.05 |
ENSDART00000162175
ENSDART00000168958 |
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr1_-_53277187 | 0.05 |
ENSDART00000131520
|
znf330
|
zinc finger protein 330 |
| chr14_+_1014109 | 0.05 |
ENSDART00000157945
|
f8
|
coagulation factor VIII, procoagulant component |
| chr17_+_19630068 | 0.04 |
ENSDART00000182619
|
rgs7a
|
regulator of G protein signaling 7a |
| chr10_-_26225548 | 0.04 |
ENSDART00000132019
ENSDART00000079194 |
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr7_+_20503344 | 0.04 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr23_-_19140781 | 0.04 |
ENSDART00000143580
|
si:ch73-381f5.2
|
si:ch73-381f5.2 |
| chr11_+_7276824 | 0.04 |
ENSDART00000185992
|
si:ch73-238c9.1
|
si:ch73-238c9.1 |
| chr6_+_49722970 | 0.04 |
ENSDART00000155934
ENSDART00000154738 |
stx16
|
syntaxin 16 |
| chr15_-_14193659 | 0.04 |
ENSDART00000171197
|
pnkp
|
polynucleotide kinase 3'-phosphatase |
| chr20_+_10723292 | 0.04 |
ENSDART00000152805
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr22_-_20386982 | 0.04 |
ENSDART00000089015
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr15_-_18115540 | 0.04 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr12_-_35582683 | 0.04 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr5_+_66220601 | 0.04 |
ENSDART00000129903
|
mfsd10
|
major facilitator superfamily domain containing 10 |
| chr1_-_44484 | 0.04 |
ENSDART00000171547
ENSDART00000164075 ENSDART00000168091 |
tmem39a
|
transmembrane protein 39A |
| chr20_-_438646 | 0.03 |
ENSDART00000009196
|
ufl1
|
UFM1-specific ligase 1 |
| chr3_-_37588855 | 0.03 |
ENSDART00000149258
|
arf2a
|
ADP-ribosylation factor 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of xbp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.5 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 1.1 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.5 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.2 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.2 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.1 | GO:1902626 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 1.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 0.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) post-translational protein modification(GO:0043687) |
| 0.0 | 0.2 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 0.1 | GO:0072672 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.1 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 1.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.2 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.7 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.5 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.4 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.3 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.5 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.2 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.0 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.0 | 0.0 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.6 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 2.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |