Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for vox
Z-value: 0.64
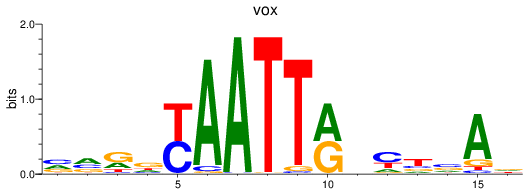
Transcription factors associated with vox
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
vox
|
ENSDARG00000099761 | ventral homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| vox | dr11_v1_chr13_-_50624173_50624173 | -0.54 | 3.5e-01 | Click! |
Activity profile of vox motif
Sorted Z-values of vox motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_54815886 | 0.76 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr18_+_8346920 | 0.38 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr21_-_32097908 | 0.35 |
ENSDART00000147387
|
si:ch211-160j14.3
|
si:ch211-160j14.3 |
| chr16_-_17197546 | 0.31 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr25_+_5035343 | 0.29 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr10_+_32104305 | 0.28 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr23_+_20110086 | 0.26 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr6_-_7720332 | 0.26 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr7_-_45999333 | 0.23 |
ENSDART00000158603
|
si:ch211-260e23.8
|
si:ch211-260e23.8 |
| chr13_+_33688474 | 0.21 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr5_+_19343880 | 0.21 |
ENSDART00000148130
|
acacb
|
acetyl-CoA carboxylase beta |
| chr7_-_23768234 | 0.21 |
ENSDART00000173981
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr22_+_16308450 | 0.20 |
ENSDART00000105678
|
lrrc39
|
leucine rich repeat containing 39 |
| chr10_+_21867307 | 0.20 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr18_+_48423973 | 0.19 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr17_+_50701748 | 0.18 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr11_+_11303458 | 0.17 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr9_+_21402863 | 0.17 |
ENSDART00000125357
|
cx30.3
|
connexin 30.3 |
| chr20_-_37813863 | 0.17 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr15_+_41027466 | 0.16 |
ENSDART00000075940
|
mtnr1ba
|
melatonin receptor type 1Ba |
| chr6_-_39649504 | 0.16 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr23_-_7674902 | 0.16 |
ENSDART00000185612
ENSDART00000180524 |
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr6_-_35401282 | 0.15 |
ENSDART00000127612
|
rgs5a
|
regulator of G protein signaling 5a |
| chr17_+_44697604 | 0.14 |
ENSDART00000156625
|
pgfb
|
placental growth factor b |
| chr2_-_48171112 | 0.14 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr5_-_29671586 | 0.14 |
ENSDART00000098336
|
spaca9
|
sperm acrosome associated 9 |
| chr21_-_17296789 | 0.13 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr20_-_49704915 | 0.13 |
ENSDART00000189232
|
COX7A2 (1 of many)
|
cytochrome c oxidase subunit 7A2 |
| chr7_+_25033924 | 0.13 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr2_-_48171441 | 0.12 |
ENSDART00000123040
|
pfkpb
|
phosphofructokinase, platelet b |
| chr5_-_42878178 | 0.12 |
ENSDART00000162981
|
CXCL11 (1 of many)
|
C-X-C motif chemokine ligand 11 |
| chr13_-_7233811 | 0.12 |
ENSDART00000162026
|
ninl
|
ninein-like |
| chr12_+_23812530 | 0.12 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr15_+_5360407 | 0.11 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr23_-_9991060 | 0.11 |
ENSDART00000111518
|
plxnb1a
|
plexin b1a |
| chr1_-_17715493 | 0.11 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr2_-_23411368 | 0.11 |
ENSDART00000159495
|
si:ch73-129a22.11
|
si:ch73-129a22.11 |
| chr5_-_25733745 | 0.11 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr22_-_7457247 | 0.11 |
ENSDART00000106081
|
BX511034.2
|
|
| chr20_+_37295006 | 0.10 |
ENSDART00000153137
|
cx23
|
connexin 23 |
| chr1_-_22691182 | 0.10 |
ENSDART00000076766
|
fgfbp2b
|
fibroblast growth factor binding protein 2b |
| chr15_+_29662401 | 0.10 |
ENSDART00000135540
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr18_+_35173683 | 0.10 |
ENSDART00000192545
|
cfap45
|
cilia and flagella associated protein 45 |
| chr9_+_21358941 | 0.10 |
ENSDART00000147619
ENSDART00000059402 |
eef1akmt1
|
EEF1A lysine methyltransferase 1 |
| chr11_-_33868881 | 0.10 |
ENSDART00000163295
ENSDART00000172633 ENSDART00000171439 |
si:ch211-227n13.3
|
si:ch211-227n13.3 |
| chr6_-_52348562 | 0.09 |
ENSDART00000142565
ENSDART00000145369 ENSDART00000016890 |
eif6
|
eukaryotic translation initiation factor 6 |
| chr17_+_17764979 | 0.09 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr21_+_39948300 | 0.09 |
ENSDART00000137740
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr18_+_1837668 | 0.09 |
ENSDART00000164210
|
CABZ01079192.1
|
|
| chr7_+_48667081 | 0.09 |
ENSDART00000083473
|
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr8_-_39822917 | 0.09 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr22_+_2198915 | 0.08 |
ENSDART00000162347
|
znf1166
|
zinc finger protein 1166 |
| chr16_+_42471455 | 0.08 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr12_-_7157527 | 0.08 |
ENSDART00000152274
|
si:dkey-187i8.2
|
si:dkey-187i8.2 |
| chr7_+_6652967 | 0.08 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr1_-_55263736 | 0.08 |
ENSDART00000152504
ENSDART00000152687 |
si:ch211-286b5.4
|
si:ch211-286b5.4 |
| chr19_+_46158078 | 0.08 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr2_-_10896745 | 0.08 |
ENSDART00000114609
|
cdcp2
|
CUB domain containing protein 2 |
| chr15_+_3790556 | 0.08 |
ENSDART00000181467
|
RNF14 (1 of many)
|
ring finger protein 14 |
| chr3_-_32337653 | 0.08 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr20_+_26966725 | 0.08 |
ENSDART00000029781
|
ahsa1a
|
AHA1, activator of heat shock protein ATPase homolog 1a |
| chr20_-_43771871 | 0.07 |
ENSDART00000153304
|
matn3a
|
matrilin 3a |
| chr10_-_35149513 | 0.07 |
ENSDART00000063434
ENSDART00000131291 |
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr24_+_19415124 | 0.07 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr21_+_15713097 | 0.07 |
ENSDART00000015841
|
gstt1b
|
glutathione S-transferase theta 1b |
| chr22_-_17474583 | 0.07 |
ENSDART00000148027
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr22_+_6905210 | 0.07 |
ENSDART00000167530
|
CABZ01065328.2
|
|
| chr5_-_64883082 | 0.07 |
ENSDART00000064983
ENSDART00000139066 |
krt1-c5
|
keratin, type 1, gene c5 |
| chr10_-_8046764 | 0.07 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr2_-_39558643 | 0.06 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr20_+_35484070 | 0.06 |
ENSDART00000026234
ENSDART00000141675 |
mep1a.2
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 2 |
| chr7_-_44957503 | 0.06 |
ENSDART00000165077
|
cdh16
|
cadherin 16, KSP-cadherin |
| chr5_+_38752287 | 0.06 |
ENSDART00000133571
|
cxcl11.8
|
chemokine (C-X-C motif) ligand 11, duplicate 8 |
| chr19_+_31541607 | 0.06 |
ENSDART00000181604
|
gmnn
|
geminin, DNA replication inhibitor |
| chr13_-_33227411 | 0.06 |
ENSDART00000057386
|
golga5
|
golgin A5 |
| chr11_-_6877973 | 0.06 |
ENSDART00000160271
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr10_+_28160265 | 0.06 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr11_+_24703108 | 0.06 |
ENSDART00000159173
|
gpr25
|
G protein-coupled receptor 25 |
| chr12_-_13729263 | 0.06 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr21_+_21812311 | 0.06 |
ENSDART00000151253
|
neu3.4
|
sialidase 3 (membrane sialidase), tandem duplicate 4 |
| chr22_-_22337382 | 0.06 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr16_-_21799407 | 0.06 |
ENSDART00000123717
|
FP015862.4
|
|
| chr15_+_20344670 | 0.06 |
ENSDART00000158986
|
plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr15_-_38154616 | 0.06 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
| chr8_-_5220125 | 0.05 |
ENSDART00000035676
|
bnip3la
|
BCL2 interacting protein 3 like a |
| chr3_-_34084387 | 0.05 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr4_+_32384950 | 0.05 |
ENSDART00000151910
|
zgc:174698
|
zgc:174698 |
| chr10_-_40968095 | 0.05 |
ENSDART00000184104
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr17_+_33311784 | 0.05 |
ENSDART00000156229
|
si:ch211-132f19.7
|
si:ch211-132f19.7 |
| chr4_-_67800414 | 0.05 |
ENSDART00000160213
|
si:ch211-66c13.1
|
si:ch211-66c13.1 |
| chr21_+_20396858 | 0.05 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr2_+_49799470 | 0.05 |
ENSDART00000146325
|
si:ch211-190k17.19
|
si:ch211-190k17.19 |
| chr2_+_33747509 | 0.05 |
ENSDART00000134187
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr4_+_35594200 | 0.05 |
ENSDART00000193902
|
si:dkeyp-4c4.2
|
si:dkeyp-4c4.2 |
| chr25_+_31868268 | 0.05 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr5_+_56268436 | 0.05 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr19_+_7735157 | 0.05 |
ENSDART00000186717
|
tuft1b
|
tuftelin 1b |
| chr12_-_19119176 | 0.05 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr2_-_2451181 | 0.05 |
ENSDART00000101033
|
pth1ra
|
parathyroid hormone 1 receptor a |
| chr4_+_32385395 | 0.04 |
ENSDART00000109176
|
zgc:174698
|
zgc:174698 |
| chr8_+_11471350 | 0.04 |
ENSDART00000092355
ENSDART00000136184 |
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr1_+_57311901 | 0.04 |
ENSDART00000149397
|
mycbpap
|
mycbp associated protein |
| chr2_-_28671139 | 0.04 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr21_-_40317035 | 0.04 |
ENSDART00000143648
ENSDART00000013359 |
or101-1
|
odorant receptor, family B, subfamily 101, member 1 |
| chr12_-_18414536 | 0.04 |
ENSDART00000132956
ENSDART00000078724 |
atpaf2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr22_+_7738966 | 0.04 |
ENSDART00000147073
|
si:ch73-44m9.5
|
si:ch73-44m9.5 |
| chr4_-_50930346 | 0.04 |
ENSDART00000184245
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr22_+_2198246 | 0.04 |
ENSDART00000165168
ENSDART00000157485 ENSDART00000168713 |
znf1157
znf1166
|
zinc finger protein 1157 zinc finger protein 1166 |
| chr1_-_11596829 | 0.04 |
ENSDART00000140725
|
si:dkey-26i13.7
|
si:dkey-26i13.7 |
| chr11_+_2391649 | 0.03 |
ENSDART00000104571
|
igfbp6a
|
insulin-like growth factor binding protein 6a |
| chr13_+_8677166 | 0.03 |
ENSDART00000181016
ENSDART00000135028 |
prop1
|
PROP paired-like homeobox 1 |
| chr6_-_39275793 | 0.03 |
ENSDART00000180477
ENSDART00000148531 |
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr17_-_33289304 | 0.03 |
ENSDART00000135118
ENSDART00000040346 |
efr3ba
|
EFR3 homolog Ba (S. cerevisiae) |
| chr8_+_11325310 | 0.03 |
ENSDART00000142577
|
fxn
|
frataxin |
| chr7_-_34262080 | 0.03 |
ENSDART00000183246
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr4_-_49119557 | 0.03 |
ENSDART00000150263
|
znf1036
|
zinc finger protein 1036 |
| chr25_-_18953322 | 0.03 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr22_-_36530902 | 0.03 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr14_-_33105434 | 0.03 |
ENSDART00000163795
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr10_+_11265387 | 0.03 |
ENSDART00000038888
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr11_+_2391469 | 0.03 |
ENSDART00000182121
|
igfbp6a
|
insulin-like growth factor binding protein 6a |
| chr21_+_6751760 | 0.03 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr25_-_21507676 | 0.03 |
ENSDART00000010706
|
immp2l
|
inner mitochondrial membrane peptidase subunit 2 |
| chr3_-_36690348 | 0.03 |
ENSDART00000192513
|
myh11b
|
myosin, heavy chain 11b, smooth muscle |
| chr23_+_41679586 | 0.03 |
ENSDART00000067662
|
CU914487.1
|
|
| chr9_-_52962521 | 0.03 |
ENSDART00000170419
|
CU855885.1
|
|
| chr3_+_16724614 | 0.03 |
ENSDART00000182135
|
gys1
|
glycogen synthase 1 (muscle) |
| chr2_+_54798689 | 0.02 |
ENSDART00000183426
|
twsg1a
|
twisted gastrulation BMP signaling modulator 1a |
| chr7_+_66048102 | 0.02 |
ENSDART00000104523
|
arntl1b
|
aryl hydrocarbon receptor nuclear translocator-like 1b |
| chr5_+_7989210 | 0.02 |
ENSDART00000168071
|
gdnfb
|
glial cell derived neurotrophic factor b |
| chr15_-_18200358 | 0.02 |
ENSDART00000158569
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr3_-_34586403 | 0.02 |
ENSDART00000151515
|
sept9a
|
septin 9a |
| chr16_-_7443388 | 0.02 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr3_-_33971297 | 0.02 |
ENSDART00000151682
|
ighj2-1
|
immunoglobulin heavy joining 2-1 |
| chr4_+_54645654 | 0.02 |
ENSDART00000192864
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr3_-_38783951 | 0.02 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr22_-_17474781 | 0.02 |
ENSDART00000186817
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr4_+_49322310 | 0.01 |
ENSDART00000184154
ENSDART00000167162 |
BX942819.1
|
|
| chr20_-_34663209 | 0.01 |
ENSDART00000132545
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr19_-_7690975 | 0.01 |
ENSDART00000151384
|
si:dkey-204a24.10
|
si:dkey-204a24.10 |
| chr4_-_41712014 | 0.01 |
ENSDART00000138165
|
znf976
|
zinc finger protein 976 |
| chr16_-_39477509 | 0.01 |
ENSDART00000191330
|
stt3b
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr5_+_28160503 | 0.01 |
ENSDART00000051516
|
tacr1a
|
tachykinin receptor 1a |
| chr7_-_59210882 | 0.01 |
ENSDART00000170330
ENSDART00000158996 |
nagk
|
N-acetylglucosamine kinase |
| chr10_-_35257458 | 0.01 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr1_+_27023687 | 0.01 |
ENSDART00000189093
|
bnc2
|
basonuclin 2 |
| chr24_+_6107901 | 0.01 |
ENSDART00000156419
|
si:ch211-37e10.2
|
si:ch211-37e10.2 |
| chr6_+_28051978 | 0.01 |
ENSDART00000143218
|
si:ch73-194h10.2
|
si:ch73-194h10.2 |
| chr16_-_29397395 | 0.01 |
ENSDART00000130757
|
tlr18
|
toll-like receptor 18 |
| chr25_+_19149241 | 0.00 |
ENSDART00000184982
ENSDART00000067324 |
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr14_+_30568961 | 0.00 |
ENSDART00000184303
|
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr19_-_10323845 | 0.00 |
ENSDART00000151259
ENSDART00000151821 |
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr16_+_20056030 | 0.00 |
ENSDART00000027020
|
ankrd28b
|
ankyrin repeat domain 28b |
| chr19_-_10971230 | 0.00 |
ENSDART00000166196
|
LO018584.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of vox
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:1902626 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.3 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.4 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.0 | 1.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) |
| 0.0 | 0.1 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.1 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.0 | GO:0060688 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.0 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 1.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.4 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.0 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.0 | GO:0016530 | metallochaperone activity(GO:0016530) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |