Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
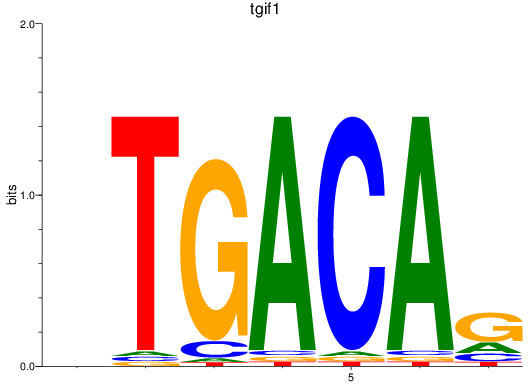
Results for tgif1
Z-value: 2.77
Transcription factors associated with tgif1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tgif1
|
ENSDARG00000059337 | TGFB-induced factor homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tgif1 | dr11_v1_chr24_-_9300160_9300160 | 0.42 | 4.8e-01 | Click! |
Activity profile of tgif1 motif
Sorted Z-values of tgif1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_36378494 | 2.41 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr15_-_4415917 | 2.01 |
ENSDART00000062874
|
atp1b3b
|
ATPase Na+/K+ transporting subunit beta 3b |
| chr4_-_815871 | 1.77 |
ENSDART00000067455
|
dpysl5b
|
dihydropyrimidinase-like 5b |
| chr20_+_34933183 | 1.74 |
ENSDART00000062738
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr19_+_43439457 | 1.62 |
ENSDART00000151571
|
ahdc1
|
AT hook, DNA binding motif, containing 1 |
| chr11_+_1796426 | 1.54 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr1_-_12278056 | 1.51 |
ENSDART00000139336
ENSDART00000137463 |
cplx2l
|
complexin 2, like |
| chr16_-_19890303 | 1.47 |
ENSDART00000147161
ENSDART00000079159 |
hdac9b
|
histone deacetylase 9b |
| chr11_-_1131569 | 1.44 |
ENSDART00000173228
|
slc6a11b
|
solute carrier family 6 (neurotransmitter transporter), member 11b |
| chr2_-_44283554 | 1.44 |
ENSDART00000184684
|
mpz
|
myelin protein zero |
| chr2_-_12502495 | 1.37 |
ENSDART00000186781
|
dpp6b
|
dipeptidyl-peptidase 6b |
| chr17_-_26926577 | 1.37 |
ENSDART00000050202
|
rcan3
|
regulator of calcineurin 3 |
| chr16_+_10776688 | 1.36 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr3_-_21137362 | 1.34 |
ENSDART00000104051
|
cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr10_-_39011514 | 1.33 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr8_-_53195500 | 1.32 |
ENSDART00000132000
|
gabrd
|
gamma-aminobutyric acid (GABA) A receptor, delta |
| chr3_-_32320537 | 1.28 |
ENSDART00000113550
ENSDART00000168483 |
si:dkey-16p21.7
|
si:dkey-16p21.7 |
| chr14_+_35691889 | 1.27 |
ENSDART00000074685
|
glrbb
|
glycine receptor, beta b |
| chr20_+_50956369 | 1.26 |
ENSDART00000170854
|
gphnb
|
gephyrin b |
| chr14_-_33297287 | 1.26 |
ENSDART00000045555
ENSDART00000138294 ENSDART00000075056 |
rab41
|
RAB41, member RAS oncogene family |
| chr18_+_13792490 | 1.26 |
ENSDART00000136754
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr1_-_45553602 | 1.24 |
ENSDART00000143664
|
grin2bb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B, genome duplicate b |
| chr16_-_13789908 | 1.23 |
ENSDART00000138540
|
ttyh1
|
tweety family member 1 |
| chr12_+_27285994 | 1.22 |
ENSDART00000087204
|
dusp3a
|
dual specificity phosphatase 3a |
| chr14_+_22172047 | 1.20 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr23_+_19558574 | 1.18 |
ENSDART00000137811
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr5_+_28433547 | 1.18 |
ENSDART00000172218
|
nsmfb
|
NMDA receptor synaptonuclear signaling and neuronal migration factor b |
| chr13_-_27660955 | 1.17 |
ENSDART00000188651
ENSDART00000134494 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr24_+_39158283 | 1.17 |
ENSDART00000053139
|
atp6v0cb
|
ATPase H+ transporting V0 subunit cb |
| chr13_-_11644806 | 1.16 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr9_-_35334642 | 1.16 |
ENSDART00000157195
|
ncam2
|
neural cell adhesion molecule 2 |
| chr19_-_20114149 | 1.15 |
ENSDART00000052620
|
npy
|
neuropeptide Y |
| chr2_+_20430366 | 1.15 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr23_+_44741500 | 1.13 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr24_-_21512425 | 1.13 |
ENSDART00000184857
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr25_-_13363286 | 1.13 |
ENSDART00000163735
ENSDART00000169119 |
ndrg4
|
NDRG family member 4 |
| chr24_-_38079261 | 1.11 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr1_-_7912932 | 1.10 |
ENSDART00000147383
|
mmd2b
|
monocyte to macrophage differentiation-associated 2b |
| chr18_+_18612388 | 1.10 |
ENSDART00000186455
|
st3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr10_-_34870667 | 1.09 |
ENSDART00000161272
|
dclk1a
|
doublecortin-like kinase 1a |
| chr11_+_25101220 | 1.07 |
ENSDART00000183700
|
ndrg3a
|
ndrg family member 3a |
| chr9_+_40939336 | 1.07 |
ENSDART00000100386
|
mstnb
|
myostatin b |
| chr18_-_21271373 | 1.07 |
ENSDART00000060001
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr8_-_39978767 | 1.06 |
ENSDART00000083066
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr6_+_38381957 | 1.04 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr18_+_783936 | 1.04 |
ENSDART00000193357
|
rpp25b
|
ribonuclease P and MRP subunit p25, b |
| chr11_-_9948487 | 1.01 |
ENSDART00000189677
ENSDART00000113171 |
nlgn1
|
neuroligin 1 |
| chr7_+_30875273 | 1.00 |
ENSDART00000173693
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr12_+_3022882 | 1.00 |
ENSDART00000122905
|
rac3b
|
Rac family small GTPase 3b |
| chr5_-_10768258 | 1.00 |
ENSDART00000157043
|
rtn4r
|
reticulon 4 receptor |
| chr1_+_7679328 | 1.00 |
ENSDART00000163488
ENSDART00000190070 |
en1b
|
engrailed homeobox 1b |
| chr17_+_47174525 | 0.99 |
ENSDART00000156831
|
CLIP4
|
si:dkeyp-47f9.4 |
| chr7_-_38633867 | 0.99 |
ENSDART00000137424
|
c1qtnf4
|
C1q and TNF related 4 |
| chr7_-_42206720 | 0.99 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr5_-_13766651 | 0.99 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr23_-_29667544 | 0.98 |
ENSDART00000059339
|
clstn1
|
calsyntenin 1 |
| chr21_+_18353703 | 0.98 |
ENSDART00000181396
ENSDART00000166359 |
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr4_-_14328997 | 0.97 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr6_+_14949950 | 0.96 |
ENSDART00000149202
ENSDART00000149949 |
pou3f3b
|
POU class 3 homeobox 3b |
| chr17_-_16965809 | 0.96 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr24_+_9744012 | 0.96 |
ENSDART00000129656
|
tmem108
|
transmembrane protein 108 |
| chr23_+_22200467 | 0.96 |
ENSDART00000025414
|
slc2a1a
|
solute carrier family 2 (facilitated glucose transporter), member 1a |
| chr19_-_30524952 | 0.96 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr23_-_30431333 | 0.95 |
ENSDART00000146633
|
camta1a
|
calmodulin binding transcription activator 1a |
| chr8_-_33114202 | 0.95 |
ENSDART00000098840
|
ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr14_-_2355833 | 0.95 |
ENSDART00000157677
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.6 |
| chr5_+_20147830 | 0.95 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr17_+_12767640 | 0.94 |
ENSDART00000112469
ENSDART00000154984 ENSDART00000124584 ENSDART00000193647 ENSDART00000179686 |
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr1_-_12278522 | 0.94 |
ENSDART00000142122
ENSDART00000003825 |
cplx2l
|
complexin 2, like |
| chr8_+_17078692 | 0.94 |
ENSDART00000023206
|
plk2b
|
polo-like kinase 2b (Drosophila) |
| chr20_-_42100932 | 0.93 |
ENSDART00000191930
|
slc35f1
|
solute carrier family 35, member F1 |
| chr15_-_15357178 | 0.93 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr6_-_14146979 | 0.93 |
ENSDART00000089564
|
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr24_+_37370064 | 0.91 |
ENSDART00000185870
|
si:ch211-183d21.3
|
si:ch211-183d21.3 |
| chr21_+_25092300 | 0.91 |
ENSDART00000168505
|
dixdc1b
|
DIX domain containing 1b |
| chr21_+_25181003 | 0.91 |
ENSDART00000169700
|
si:dkey-183i3.9
|
si:dkey-183i3.9 |
| chr9_+_42066030 | 0.91 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr11_-_762721 | 0.90 |
ENSDART00000166465
|
syn2b
|
synapsin IIb |
| chr17_-_43287290 | 0.90 |
ENSDART00000156885
|
EML5
|
si:dkey-1f12.3 |
| chr6_-_24358732 | 0.89 |
ENSDART00000159595
|
ephx4
|
epoxide hydrolase 4 |
| chr21_-_12119711 | 0.89 |
ENSDART00000131538
|
celf4
|
CUGBP, Elav-like family member 4 |
| chr8_-_14375890 | 0.89 |
ENSDART00000090306
|
xpr1a
|
xenotropic and polytropic retrovirus receptor 1a |
| chr12_+_18578597 | 0.89 |
ENSDART00000134944
|
grid2ipb
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b |
| chr13_-_50672341 | 0.88 |
ENSDART00000190498
|
FP102122.1
|
|
| chr11_-_1509773 | 0.87 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr24_-_29997145 | 0.87 |
ENSDART00000135094
|
palmdb
|
palmdelphin b |
| chr8_-_17067364 | 0.87 |
ENSDART00000132687
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr2_-_24444838 | 0.87 |
ENSDART00000147885
ENSDART00000164720 |
kcnn1a
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1a |
| chr3_+_58133227 | 0.85 |
ENSDART00000154566
|
si:ch211-256e16.3
|
si:ch211-256e16.3 |
| chr3_-_28428198 | 0.85 |
ENSDART00000151546
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr5_+_39504136 | 0.85 |
ENSDART00000121460
|
prdm8b
|
PR domain containing 8b |
| chr10_-_22845485 | 0.85 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr3_+_45401472 | 0.84 |
ENSDART00000156693
|
baiap3
|
BAI1-associated protein 3 |
| chr17_-_17130942 | 0.84 |
ENSDART00000064241
|
nrxn3a
|
neurexin 3a |
| chr12_-_25916530 | 0.83 |
ENSDART00000186386
|
sncgb
|
synuclein, gamma b (breast cancer-specific protein 1) |
| chr3_+_54047342 | 0.83 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr9_+_17862858 | 0.83 |
ENSDART00000166566
|
dgkh
|
diacylglycerol kinase, eta |
| chr16_-_13730152 | 0.83 |
ENSDART00000138772
|
ttyh1
|
tweety family member 1 |
| chr19_-_874888 | 0.82 |
ENSDART00000007206
|
eomesa
|
eomesodermin homolog a |
| chr11_-_37509001 | 0.82 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr21_+_25643880 | 0.81 |
ENSDART00000192392
ENSDART00000145091 |
tmem151a
|
transmembrane protein 151A |
| chr25_+_19999623 | 0.81 |
ENSDART00000026401
|
zgc:194665
|
zgc:194665 |
| chr8_+_8845932 | 0.81 |
ENSDART00000112028
|
si:ch211-180f4.1
|
si:ch211-180f4.1 |
| chr21_-_43040780 | 0.81 |
ENSDART00000187037
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr7_+_42206543 | 0.81 |
ENSDART00000112543
|
phkb
|
phosphorylase kinase, beta |
| chr11_+_37275448 | 0.80 |
ENSDART00000161423
|
creld1a
|
cysteine-rich with EGF-like domains 1a |
| chr10_+_22381802 | 0.80 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr23_-_29505463 | 0.79 |
ENSDART00000050915
|
kif1b
|
kinesin family member 1B |
| chr4_+_4803698 | 0.79 |
ENSDART00000129252
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr11_-_44163164 | 0.79 |
ENSDART00000047126
|
clcn4
|
chloride channel, voltage-sensitive 4 |
| chr5_-_21422390 | 0.79 |
ENSDART00000144198
|
tenm1
|
teneurin transmembrane protein 1 |
| chr17_-_43466317 | 0.79 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr6_+_38427570 | 0.79 |
ENSDART00000170612
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr12_-_10275320 | 0.79 |
ENSDART00000170078
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr25_+_33939728 | 0.78 |
ENSDART00000148537
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr5_+_54685175 | 0.78 |
ENSDART00000115016
|
pmchl
|
pro-melanin-concentrating hormone, like |
| chr11_+_28218141 | 0.78 |
ENSDART00000043756
|
ephb2b
|
eph receptor B2b |
| chr7_-_69636502 | 0.78 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr25_-_3979288 | 0.77 |
ENSDART00000157117
|
myrf
|
myelin regulatory factor |
| chr15_+_47161917 | 0.77 |
ENSDART00000167860
|
gap43
|
growth associated protein 43 |
| chr1_+_12766351 | 0.77 |
ENSDART00000165785
|
pcdh10a
|
protocadherin 10a |
| chr20_+_19238382 | 0.77 |
ENSDART00000136757
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr9_-_3653259 | 0.77 |
ENSDART00000140425
ENSDART00000025332 |
gad1a
|
glutamate decarboxylase 1a |
| chr5_+_27137473 | 0.77 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr9_-_30336125 | 0.76 |
ENSDART00000131683
|
sytl5
|
synaptotagmin-like 5 |
| chr24_+_4978055 | 0.76 |
ENSDART00000045813
|
zic4
|
zic family member 4 |
| chr23_+_40460333 | 0.76 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr16_+_11483811 | 0.76 |
ENSDART00000169012
ENSDART00000173042 |
grik5
|
glutamate receptor, ionotropic, kainate 5 |
| chr3_+_46459540 | 0.75 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr16_+_34528409 | 0.75 |
ENSDART00000144718
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr9_-_31278048 | 0.75 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr15_-_9272328 | 0.75 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr23_+_21663631 | 0.75 |
ENSDART00000066125
|
dhrs3a
|
dehydrogenase/reductase (SDR family) member 3a |
| chr5_-_51830997 | 0.75 |
ENSDART00000163616
|
homer1b
|
homer scaffolding protein 1b |
| chr12_+_7491690 | 0.75 |
ENSDART00000152564
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr16_-_25533413 | 0.74 |
ENSDART00000145825
|
dtnbp1a
|
dystrobrevin binding protein 1a |
| chr12_+_29055143 | 0.74 |
ENSDART00000076322
|
gabrz
|
gamma-aminobutyric acid (GABA) A receptor, zeta |
| chr7_-_58269812 | 0.74 |
ENSDART00000050077
|
sdcbp
|
syndecan binding protein (syntenin) |
| chr5_+_70155935 | 0.74 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr4_-_19016396 | 0.74 |
ENSDART00000166160
|
si:dkey-31f5.11
|
si:dkey-31f5.11 |
| chr20_+_50852356 | 0.74 |
ENSDART00000167517
ENSDART00000168396 |
gphnb
|
gephyrin b |
| chr8_+_39619087 | 0.74 |
ENSDART00000134822
|
msi1
|
musashi RNA-binding protein 1 |
| chr11_-_43226255 | 0.73 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr19_-_15397946 | 0.73 |
ENSDART00000143480
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr9_+_23511913 | 0.73 |
ENSDART00000137759
|
cntnap5a
|
contactin associated protein-like 5a |
| chr8_+_50727220 | 0.73 |
ENSDART00000127062
|
egr3
|
early growth response 3 |
| chr11_-_10770053 | 0.73 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr7_-_55675617 | 0.73 |
ENSDART00000021009
ENSDART00000188631 |
cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr9_+_23255410 | 0.73 |
ENSDART00000113241
ENSDART00000137231 |
tmem163a
|
transmembrane protein 163a |
| chr1_+_41666611 | 0.73 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr12_-_4781801 | 0.73 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr13_-_23756700 | 0.72 |
ENSDART00000057612
|
rgs17
|
regulator of G protein signaling 17 |
| chr13_+_2448251 | 0.72 |
ENSDART00000188361
|
arfgef3
|
ARFGEF family member 3 |
| chr10_-_32890617 | 0.72 |
ENSDART00000134922
|
kctd7
|
potassium channel tetramerization domain containing 7 |
| chr10_+_34426256 | 0.72 |
ENSDART00000102566
|
nbeaa
|
neurobeachin a |
| chr18_-_26126258 | 0.72 |
ENSDART00000145875
|
ANKRD34C
|
ankyrin repeat domain 34C |
| chr13_-_36545258 | 0.72 |
ENSDART00000186171
|
FP103009.1
|
|
| chr13_-_36911118 | 0.71 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr24_-_20531813 | 0.71 |
ENSDART00000089428
|
ctdspla
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like a |
| chr7_+_73332486 | 0.71 |
ENSDART00000174119
ENSDART00000092051 ENSDART00000192388 |
CABZ01081780.1
|
|
| chr8_-_17771755 | 0.71 |
ENSDART00000063592
|
prkcz
|
protein kinase C, zeta |
| chr10_+_22782522 | 0.71 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr19_-_9712530 | 0.71 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr16_-_23372666 | 0.71 |
ENSDART00000158786
|
trim46a
|
tripartite motif containing 46a |
| chr9_-_9348058 | 0.71 |
ENSDART00000132257
|
zgc:113337
|
zgc:113337 |
| chr9_+_22634073 | 0.71 |
ENSDART00000181822
|
etv5a
|
ets variant 5a |
| chr13_+_2732951 | 0.71 |
ENSDART00000136977
ENSDART00000114328 |
wdr11
|
WD repeat domain 11 |
| chr5_-_45877387 | 0.70 |
ENSDART00000183714
ENSDART00000041503 |
slc4a4a
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4a |
| chr16_+_41873708 | 0.69 |
ENSDART00000084631
ENSDART00000084639 ENSDART00000058611 |
scn1ba
|
sodium channel, voltage-gated, type I, beta a |
| chr24_-_35767501 | 0.69 |
ENSDART00000105680
ENSDART00000042290 ENSDART00000166264 |
dtna
|
dystrobrevin, alpha |
| chr3_-_22216442 | 0.69 |
ENSDART00000153613
|
maptb
|
microtubule-associated protein tau b |
| chr21_-_11054876 | 0.69 |
ENSDART00000146576
|
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr22_+_5574952 | 0.68 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr25_-_4482449 | 0.68 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr7_-_34329527 | 0.68 |
ENSDART00000173454
|
madd
|
MAP-kinase activating death domain |
| chr7_-_19600181 | 0.68 |
ENSDART00000100757
|
oxa1l
|
oxidase (cytochrome c) assembly 1-like |
| chr19_-_31686252 | 0.68 |
ENSDART00000131721
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr1_+_45085194 | 0.68 |
ENSDART00000193863
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr20_-_44576949 | 0.68 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr7_-_72067475 | 0.68 |
ENSDART00000017763
|
LO018380.1
|
|
| chr19_-_19599372 | 0.67 |
ENSDART00000160002
ENSDART00000171664 |
tax1bp1a
|
Tax1 (human T-cell leukemia virus type I) binding protein 1a |
| chr10_-_24371312 | 0.67 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr24_+_4977862 | 0.67 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr9_-_44642108 | 0.67 |
ENSDART00000086202
|
pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr12_-_4672708 | 0.67 |
ENSDART00000152659
ENSDART00000186076 |
si:ch211-255p10.4
|
si:ch211-255p10.4 |
| chr19_+_37848830 | 0.67 |
ENSDART00000042276
ENSDART00000180872 |
nxph1
|
neurexophilin 1 |
| chr3_-_22216771 | 0.67 |
ENSDART00000130546
|
maptb
|
microtubule-associated protein tau b |
| chr20_+_26095530 | 0.67 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr5_+_24245682 | 0.66 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr18_+_27738349 | 0.66 |
ENSDART00000187816
|
tspan18b
|
tetraspanin 18b |
| chr2_-_44038698 | 0.65 |
ENSDART00000079582
ENSDART00000146804 |
kirrel1b
|
kirre like nephrin family adhesion molecule 1b |
| chr13_+_28702104 | 0.65 |
ENSDART00000135481
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr2_-_10062575 | 0.65 |
ENSDART00000091726
|
fam78ba
|
family with sequence similarity 78, member B a |
| chr7_+_39758714 | 0.65 |
ENSDART00000111278
|
sorcs2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr5_-_41307550 | 0.65 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr21_-_27010796 | 0.65 |
ENSDART00000065398
ENSDART00000144342 ENSDART00000126542 |
ppp1r14ba
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ba |
| chr5_+_64319590 | 0.65 |
ENSDART00000192652
|
FQ377918.1
|
|
| chr16_-_29164379 | 0.64 |
ENSDART00000132589
|
mef2d
|
myocyte enhancer factor 2d |
| chr2_+_21982911 | 0.64 |
ENSDART00000190722
ENSDART00000044371 ENSDART00000134912 |
tox
|
thymocyte selection-associated high mobility group box |
| chr3_-_19133003 | 0.64 |
ENSDART00000145215
|
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
Network of associatons between targets according to the STRING database.
First level regulatory network of tgif1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0018315 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.3 | 1.3 | GO:0071788 | endoplasmic reticulum tubular network maintenance(GO:0071788) |
| 0.3 | 1.1 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.3 | 0.3 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.3 | 0.8 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.3 | 0.8 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.2 | 1.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 4.9 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.2 | 0.7 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.2 | 0.7 | GO:2000402 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.2 | 0.9 | GO:0045843 | negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle tissue development(GO:1901862) |
| 0.2 | 1.4 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.2 | 0.7 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.2 | 0.6 | GO:0097401 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.2 | 1.3 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.2 | 1.3 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.2 | 0.9 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.7 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.1 | 1.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.9 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.1 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.1 | 0.7 | GO:0032979 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.4 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.1 | 1.6 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 0.5 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.1 | 1.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.8 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.1 | 0.4 | GO:0099623 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 | 0.5 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.1 | 0.6 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.5 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 1.0 | GO:0046323 | glucose import(GO:0046323) |
| 0.1 | 0.5 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 1.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 1.2 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 1.0 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.8 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 1.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.3 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.1 | 0.3 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.5 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 0.5 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 1.4 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.8 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.6 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 1.9 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 1.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.4 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.1 | 1.1 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.4 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 1.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.5 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 0.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.7 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.1 | 0.6 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.3 | GO:0015824 | L-alanine transport(GO:0015808) proline transport(GO:0015824) |
| 0.1 | 0.7 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 1.0 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.4 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.5 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.4 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.4 | GO:0003413 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.1 | 0.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 0.6 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.5 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.6 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.1 | 0.4 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 1.8 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.1 | 0.5 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 1.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.2 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.7 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 0.6 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.5 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.2 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.4 | GO:0072160 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.1 | 1.1 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.4 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.2 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.1 | 0.9 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.1 | 0.4 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.3 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.1 | 1.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.8 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 2.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.6 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.5 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.3 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.1 | 1.0 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.1 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 2.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 0.4 | GO:0043901 | negative regulation of multi-organism process(GO:0043901) |
| 0.1 | 1.8 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 0.6 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.1 | 0.2 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 0.3 | GO:0006660 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.3 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.5 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.6 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.2 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 2.2 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 7.6 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 1.7 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.5 | GO:0043217 | myelin maintenance(GO:0043217) |
| 0.1 | 0.9 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.3 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.4 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.1 | 0.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 0.4 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 0.2 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.1 | 0.3 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.2 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.1 | 0.5 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.2 | GO:0044107 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.1 | 1.2 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.1 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.5 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.4 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.2 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 2.0 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.2 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.1 | 0.4 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.4 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 1.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.6 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.9 | GO:0072176 | pronephric duct development(GO:0039022) nephric duct development(GO:0072176) |
| 0.0 | 0.2 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.3 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 1.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 1.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.1 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 1.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.4 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.1 | GO:0060945 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.0 | 1.9 | GO:0021854 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 1.4 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.6 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.8 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.6 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.2 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.0 | 0.3 | GO:0031116 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.3 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.5 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.2 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.2 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.2 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.0 | 0.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.6 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.8 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.3 | GO:0046661 | male sex differentiation(GO:0046661) |
| 0.0 | 0.4 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.2 | GO:0071698 | olfactory placode development(GO:0071698) |
| 0.0 | 0.5 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.2 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 1.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.7 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 1.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.7 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.6 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 1.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 2.2 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.2 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.4 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.0 | 0.4 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 0.9 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.4 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.6 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.8 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.0 | 0.3 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.2 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.7 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.8 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.2 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0045041 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.0 | GO:1904375 | regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.0 | 0.8 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 0.2 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 4.4 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.4 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 1.0 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.7 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.5 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.6 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 2.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.3 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.1 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.5 | GO:0051283 | sequestering of calcium ion(GO:0051208) release of sequestered calcium ion into cytosol(GO:0051209) regulation of sequestering of calcium ion(GO:0051282) negative regulation of sequestering of calcium ion(GO:0051283) calcium ion transmembrane import into cytosol(GO:0097553) calcium ion import into cytosol(GO:1902656) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.6 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.3 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.7 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.1 | GO:0097037 | heme export(GO:0097037) |
| 0.0 | 2.7 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.0 | 0.1 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.3 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 1.1 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.4 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0042551 | neuron maturation(GO:0042551) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.4 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.5 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.2 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.3 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 1.0 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.1 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.2 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 1.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.0 | 0.1 | GO:0080009 | 7-methylguanosine mRNA capping(GO:0006370) mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.0 | 0.5 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.5 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.3 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.3 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.3 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.6 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 1.1 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.1 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.3 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.9 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.7 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.0 | GO:0032371 | regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.0 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.5 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 1.0 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.0 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.0 | 0.5 | GO:0051051 | negative regulation of transport(GO:0051051) |
| 0.0 | 0.3 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0042593 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 0.3 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 1.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.4 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.1 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.9 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 1.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.0 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 0.3 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.1 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.2 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.2 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 3.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.3 | 1.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.3 | 0.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 2.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 0.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 2.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 0.7 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 0.9 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.4 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 3.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.6 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.4 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.7 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.1 | 4.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 1.2 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 0.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.4 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.2 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 1.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.4 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 1.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 1.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.7 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.6 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 1.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.4 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.7 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.2 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.8 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.3 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.4 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.8 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.4 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 2.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 3.0 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.6 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.3 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 7.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 4.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 13.5 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.8 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.1 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.0 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.4 | 2.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 0.9 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.3 | 2.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 0.8 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.2 | 0.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 1.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 1.1 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.2 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 1.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.2 | 1.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 0.2 | GO:0035004 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.2 | 1.4 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 0.5 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 1.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 0.5 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 2.1 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 0.1 | 1.8 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 2.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 4.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.5 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 1.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.4 | GO:0015222 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 1.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.4 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 4.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.6 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.4 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.1 | 0.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 1.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.5 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 1.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.4 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.5 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 0.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.3 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.3 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.1 | 0.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.5 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.5 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.4 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 1.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 1.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.7 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.5 | GO:0017136 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.1 | 0.4 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.4 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 1.2 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.5 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.6 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.3 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.2 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.1 | 1.1 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.2 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 0.1 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 1.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 1.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.4 | GO:0015154 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.5 | GO:0052659 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.1 | 0.3 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.9 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 2.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.5 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 1.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.0 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.3 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.2 | GO:0072570 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.6 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 1.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 1.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 1.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 3.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0008200 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.0 | 1.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.3 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.1 | GO:0017113 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.5 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.8 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.4 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.1 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.4 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 1.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.2 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 1.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.3 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.1 | GO:1990931 | RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 3.0 | GO:0019900 | kinase binding(GO:0019900) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.0 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 3.4 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.0 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.3 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 3.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 2.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.9 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 1.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |