Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
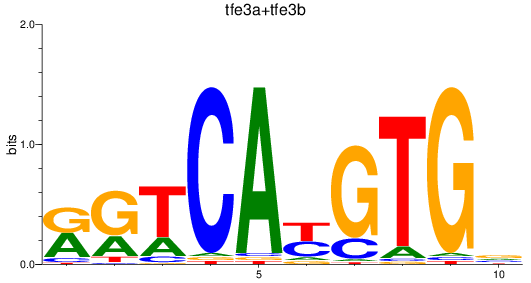
Results for tfe3a+tfe3b
Z-value: 1.76
Transcription factors associated with tfe3a+tfe3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfe3b
|
ENSDARG00000019457 | transcription factor binding to IGHM enhancer 3b |
|
tfe3a
|
ENSDARG00000098903 | transcription factor binding to IGHM enhancer 3a |
|
tfe3b
|
ENSDARG00000111231 | transcription factor binding to IGHM enhancer 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfe3b | dr11_v1_chr11_+_25504215_25504215 | -0.55 | 3.3e-01 | Click! |
| tfe3a | dr11_v1_chr8_+_7778770_7778770 | -0.44 | 4.5e-01 | Click! |
Activity profile of tfe3a+tfe3b motif
Sorted Z-values of tfe3a+tfe3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_31665836 | 1.55 |
ENSDART00000135411
ENSDART00000143914 |
si:ch211-106h4.12
|
si:ch211-106h4.12 |
| chr22_-_17677947 | 1.35 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr5_-_29534748 | 1.32 |
ENSDART00000159587
|
AL831768.1
|
|
| chr20_-_17041025 | 1.24 |
ENSDART00000063764
|
si:dkey-5n18.1
|
si:dkey-5n18.1 |
| chr16_-_45178430 | 1.22 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr3_-_16784280 | 1.19 |
ENSDART00000137108
ENSDART00000137276 |
si:dkey-30j10.5
|
si:dkey-30j10.5 |
| chr9_-_56272465 | 1.07 |
ENSDART00000039235
|
lcp1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr9_-_33063083 | 1.05 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr21_-_22673758 | 1.00 |
ENSDART00000164910
|
gig2i
|
grass carp reovirus (GCRV)-induced gene 2i |
| chr20_+_38257766 | 0.98 |
ENSDART00000147485
ENSDART00000149160 |
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr12_+_20336070 | 0.97 |
ENSDART00000066385
|
zgc:163057
|
zgc:163057 |
| chr23_-_31913231 | 0.97 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr14_+_52571134 | 0.93 |
ENSDART00000166708
|
rpl26
|
ribosomal protein L26 |
| chr20_-_39735952 | 0.92 |
ENSDART00000101049
ENSDART00000137485 ENSDART00000062402 |
tpd52l1
|
tumor protein D52-like 1 |
| chr7_-_19923249 | 0.92 |
ENSDART00000078694
|
zgc:110591
|
zgc:110591 |
| chr5_+_27404946 | 0.91 |
ENSDART00000121886
ENSDART00000005025 |
hdr
|
hematopoietic death receptor |
| chr7_-_67214972 | 0.90 |
ENSDART00000156861
|
swap70a
|
switching B cell complex subunit SWAP70a |
| chr20_-_37813863 | 0.87 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr14_-_15171435 | 0.86 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr2_+_36007449 | 0.86 |
ENSDART00000161837
|
lamc2
|
laminin, gamma 2 |
| chr2_+_24507770 | 0.85 |
ENSDART00000154802
ENSDART00000052063 |
rps28
|
ribosomal protein S28 |
| chr12_-_13155653 | 0.85 |
ENSDART00000152467
|
cxl34c
|
CX chemokine ligand 34c |
| chr18_-_1414760 | 0.83 |
ENSDART00000171881
|
PEPD
|
peptidase D |
| chr10_+_41159241 | 0.82 |
ENSDART00000141657
|
anxa4
|
annexin A4 |
| chr7_-_8374950 | 0.82 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr19_+_791538 | 0.80 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr1_+_12135129 | 0.77 |
ENSDART00000126020
|
spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr21_-_22689805 | 0.77 |
ENSDART00000157560
ENSDART00000110792 |
gig2e
|
grass carp reovirus (GCRV)-induced gene 2e |
| chr21_+_25226558 | 0.76 |
ENSDART00000168480
|
sycn.2
|
syncollin, tandem duplicate 2 |
| chr1_-_58059134 | 0.76 |
ENSDART00000160970
|
caspb
|
caspase b |
| chr18_+_44703343 | 0.75 |
ENSDART00000131510
|
b3gnt2l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2, like |
| chr19_+_48117995 | 0.74 |
ENSDART00000170865
|
nme2b.1
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 |
| chr22_+_5120033 | 0.73 |
ENSDART00000169200
|
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr23_-_5719453 | 0.72 |
ENSDART00000033093
|
lad1
|
ladinin |
| chr23_+_39089574 | 0.72 |
ENSDART00000164711
|
nfatc2a
|
nuclear factor of activated T cells 2a |
| chr25_+_10547228 | 0.71 |
ENSDART00000067678
|
zgc:110339
|
zgc:110339 |
| chr13_-_214122 | 0.71 |
ENSDART00000169273
|
ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr18_-_5509616 | 0.71 |
ENSDART00000142945
|
bloc1s6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr5_+_4564233 | 0.70 |
ENSDART00000193435
|
CABZ01058647.1
|
|
| chr21_-_42876565 | 0.68 |
ENSDART00000126480
|
zmp:0000001268
|
zmp:0000001268 |
| chr11_+_24729346 | 0.68 |
ENSDART00000087740
|
zgc:153953
|
zgc:153953 |
| chr21_-_25741096 | 0.68 |
ENSDART00000181756
|
cldnh
|
claudin h |
| chr18_+_26422124 | 0.67 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr21_-_10773344 | 0.67 |
ENSDART00000063244
|
grp
|
gastrin-releasing peptide |
| chr15_-_17099560 | 0.67 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr7_-_55633475 | 0.67 |
ENSDART00000149478
|
galns
|
galactosamine (N-acetyl)-6-sulfatase |
| chr6_+_112579 | 0.66 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr9_+_35077546 | 0.66 |
ENSDART00000142688
|
si:dkey-192g7.3
|
si:dkey-192g7.3 |
| chr3_-_39488482 | 0.66 |
ENSDART00000135192
|
zgc:100868
|
zgc:100868 |
| chr3_-_39488639 | 0.66 |
ENSDART00000161644
|
zgc:100868
|
zgc:100868 |
| chr24_+_7631797 | 0.65 |
ENSDART00000187464
|
cavin1b
|
caveolae associated protein 1b |
| chr1_+_57145072 | 0.63 |
ENSDART00000152776
|
si:ch73-94k4.4
|
si:ch73-94k4.4 |
| chr15_+_20403903 | 0.62 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr16_-_5143124 | 0.61 |
ENSDART00000131876
ENSDART00000060630 |
ttk
|
ttk protein kinase |
| chr5_+_4575800 | 0.61 |
ENSDART00000180952
|
cst14b.2
|
cystatin 14b, tandem duplicate 2 |
| chr5_-_1963498 | 0.60 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr23_+_31913292 | 0.59 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr8_+_13389115 | 0.59 |
ENSDART00000184428
ENSDART00000154266 ENSDART00000049469 |
jak3
|
Janus kinase 3 (a protein tyrosine kinase, leukocyte) |
| chr7_+_39410180 | 0.58 |
ENSDART00000168641
|
CT030188.1
|
|
| chr3_-_13546610 | 0.57 |
ENSDART00000159647
|
amdhd2
|
amidohydrolase domain containing 2 |
| chr19_-_15229421 | 0.57 |
ENSDART00000055619
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr10_-_35149513 | 0.57 |
ENSDART00000063434
ENSDART00000131291 |
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr25_-_19608382 | 0.56 |
ENSDART00000022279
ENSDART00000135201 ENSDART00000147223 ENSDART00000190220 ENSDART00000184242 ENSDART00000166824 |
gtse1
|
G-2 and S-phase expressed 1 |
| chr10_-_21953643 | 0.56 |
ENSDART00000188921
ENSDART00000193569 |
FO744833.2
|
|
| chr22_-_17653143 | 0.56 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr7_+_46019780 | 0.56 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr5_+_61475451 | 0.56 |
ENSDART00000163444
|
lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr2_-_24069331 | 0.55 |
ENSDART00000156972
ENSDART00000181691 ENSDART00000157041 |
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr16_+_20895904 | 0.55 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr21_-_22715297 | 0.55 |
ENSDART00000065548
|
c1qb
|
complement component 1, q subcomponent, B chain |
| chr24_+_42149453 | 0.55 |
ENSDART00000128766
|
serpinb1l3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 3 |
| chr19_+_7895086 | 0.55 |
ENSDART00000132180
|
si:dkey-266f7.1
|
si:dkey-266f7.1 |
| chr24_+_27511734 | 0.54 |
ENSDART00000105771
|
cxcl32b.1
|
chemokine (C-X-C motif) ligand 32b, duplicate 1 |
| chr7_-_11383933 | 0.54 |
ENSDART00000163949
|
mesd
|
mesoderm development LRP chaperone |
| chr17_-_24916889 | 0.54 |
ENSDART00000156157
|
si:ch211-195o20.7
|
si:ch211-195o20.7 |
| chr7_-_11384279 | 0.54 |
ENSDART00000102515
ENSDART00000172796 |
mesd
|
mesoderm development LRP chaperone |
| chr3_-_18805225 | 0.54 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr7_+_39410393 | 0.53 |
ENSDART00000158561
ENSDART00000185173 |
CT030188.1
|
|
| chr15_-_4580763 | 0.52 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr6_+_392815 | 0.52 |
ENSDART00000163142
|
cyth4b
|
cytohesin 4b |
| chr22_+_978867 | 0.52 |
ENSDART00000148981
|
def6b
|
differentially expressed in FDCP 6b homolog (mouse) |
| chr18_+_44649804 | 0.52 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr21_-_3007412 | 0.52 |
ENSDART00000190839
|
CKS2
|
zgc:86839 |
| chr24_-_42072886 | 0.52 |
ENSDART00000171389
|
CABZ01095370.1
|
|
| chr3_-_4591643 | 0.52 |
ENSDART00000138144
|
ftr50
|
finTRIM family, member 50 |
| chr3_+_31662126 | 0.52 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr22_+_29991834 | 0.51 |
ENSDART00000147728
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr12_+_37401331 | 0.51 |
ENSDART00000125040
|
si:ch211-152f22.4
|
si:ch211-152f22.4 |
| chr8_+_8671229 | 0.51 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr1_+_59538755 | 0.51 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr19_+_19756425 | 0.51 |
ENSDART00000167606
|
hoxa3a
|
homeobox A3a |
| chr3_-_3209432 | 0.51 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr13_+_22717366 | 0.50 |
ENSDART00000134122
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr23_+_32028574 | 0.50 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr25_+_19718504 | 0.50 |
ENSDART00000036702
|
mxd
|
myxovirus (influenza virus) resistance D |
| chr17_+_25331576 | 0.50 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr3_+_19621034 | 0.49 |
ENSDART00000025358
|
itgb3a
|
integrin beta 3a |
| chr15_-_37850969 | 0.49 |
ENSDART00000031418
|
hsc70
|
heat shock cognate 70 |
| chr2_-_10877765 | 0.49 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr7_-_59054322 | 0.49 |
ENSDART00000165390
|
chmp5b
|
charged multivesicular body protein 5b |
| chr15_+_1534644 | 0.49 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr14_+_45471642 | 0.49 |
ENSDART00000126979
ENSDART00000172952 ENSDART00000173284 |
ubxn1
|
UBX domain protein 1 |
| chr20_-_32045057 | 0.48 |
ENSDART00000152970
ENSDART00000034248 |
rab32a
|
RAB32a, member RAS oncogene family |
| chr19_-_11336782 | 0.48 |
ENSDART00000131014
|
sept7a
|
septin 7a |
| chr21_-_22669669 | 0.48 |
ENSDART00000101784
|
gig2j
|
grass carp reovirus (GCRV)-induced gene 2j |
| chr10_-_15053507 | 0.48 |
ENSDART00000157446
ENSDART00000170441 |
CLDN23
|
si:ch211-95j8.5 |
| chr1_+_14253118 | 0.48 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr11_+_10541258 | 0.47 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr15_-_28596507 | 0.47 |
ENSDART00000156800
|
si:ch211-225b7.5
|
si:ch211-225b7.5 |
| chr5_+_26204561 | 0.47 |
ENSDART00000137178
|
marveld2b
|
MARVEL domain containing 2b |
| chr25_+_245438 | 0.46 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr16_+_33143503 | 0.46 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr21_-_3700334 | 0.46 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr22_+_19494996 | 0.45 |
ENSDART00000140829
ENSDART00000135168 |
si:dkey-78l4.10
|
si:dkey-78l4.10 |
| chr7_-_73717082 | 0.45 |
ENSDART00000164301
ENSDART00000082625 |
BX664721.2
|
|
| chr23_+_44644911 | 0.45 |
ENSDART00000140799
|
zgc:85858
|
zgc:85858 |
| chr3_+_16922226 | 0.45 |
ENSDART00000017646
|
atp6v0a1a
|
ATPase H+ transporting V0 subunit a1a |
| chr7_-_6340998 | 0.45 |
ENSDART00000172884
|
zgc:165555
|
zgc:165555 |
| chr17_-_114121 | 0.45 |
ENSDART00000172408
ENSDART00000157784 |
arhgap11a
|
Rho GTPase activating protein 11A |
| chr9_+_563547 | 0.45 |
ENSDART00000162761
|
CU984600.2
|
|
| chr14_+_23709543 | 0.44 |
ENSDART00000136909
|
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr6_-_15641686 | 0.44 |
ENSDART00000135583
|
mlpha
|
melanophilin a |
| chr11_-_270210 | 0.44 |
ENSDART00000005217
ENSDART00000172779 |
alas1
|
aminolevulinate, delta-, synthase 1 |
| chr23_+_35847200 | 0.44 |
ENSDART00000129222
|
rarga
|
retinoic acid receptor gamma a |
| chr22_+_19478140 | 0.44 |
ENSDART00000135291
ENSDART00000136576 |
si:dkey-78l4.8
|
si:dkey-78l4.8 |
| chr15_-_1198886 | 0.44 |
ENSDART00000063285
|
lxn
|
latexin |
| chr19_-_808265 | 0.44 |
ENSDART00000082454
|
glmp
|
glycosylated lysosomal membrane protein |
| chr7_+_49654588 | 0.44 |
ENSDART00000025451
ENSDART00000141934 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr7_+_53156810 | 0.43 |
ENSDART00000189816
|
cdh29
|
cadherin 29 |
| chr17_-_20143946 | 0.43 |
ENSDART00000138911
|
actn2b
|
actinin, alpha 2b |
| chr7_-_5316901 | 0.43 |
ENSDART00000181505
ENSDART00000124367 |
si:cabz01074946.1
|
si:cabz01074946.1 |
| chr6_-_1761256 | 0.43 |
ENSDART00000171313
|
orc4
|
origin recognition complex, subunit 4 |
| chr18_-_11729 | 0.43 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr8_+_41229233 | 0.42 |
ENSDART00000131135
|
zgc:152830
|
zgc:152830 |
| chr17_+_53425037 | 0.42 |
ENSDART00000154983
|
fabp10b
|
fatty acid binding protein 10b, liver basic |
| chr8_+_30797363 | 0.42 |
ENSDART00000077280
|
mmp11a
|
matrix metallopeptidase 11a |
| chr7_+_49695904 | 0.42 |
ENSDART00000183550
ENSDART00000126991 |
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr8_+_17167876 | 0.41 |
ENSDART00000134665
|
cenph
|
centromere protein H |
| chr15_-_43625549 | 0.41 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr23_+_384850 | 0.41 |
ENSDART00000114000
|
zgc:101663
|
zgc:101663 |
| chr5_-_69944084 | 0.41 |
ENSDART00000188557
ENSDART00000127782 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr17_+_53424415 | 0.41 |
ENSDART00000157022
|
slc9a1b
|
solute carrier family 9 member A1b |
| chr23_+_32039386 | 0.40 |
ENSDART00000133801
|
mylk2
|
myosin light chain kinase 2 |
| chr7_-_3669868 | 0.40 |
ENSDART00000172907
|
si:ch211-282j17.12
|
si:ch211-282j17.12 |
| chr20_+_7584211 | 0.40 |
ENSDART00000132481
ENSDART00000127975 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr10_+_36650222 | 0.40 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr7_-_3693303 | 0.40 |
ENSDART00000166949
|
si:ch211-282j17.13
|
si:ch211-282j17.13 |
| chr10_+_36662640 | 0.40 |
ENSDART00000063359
|
ucp2
|
uncoupling protein 2 |
| chr9_+_36946340 | 0.39 |
ENSDART00000135281
|
si:dkey-3d4.3
|
si:dkey-3d4.3 |
| chr19_+_7152966 | 0.39 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr7_-_15118140 | 0.39 |
ENSDART00000179985
|
si:dkey-172h23.2
|
si:dkey-172h23.2 |
| chr10_+_32050906 | 0.38 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr11_-_42730063 | 0.38 |
ENSDART00000169776
|
si:ch73-106k19.2
|
si:ch73-106k19.2 |
| chr3_+_43086548 | 0.38 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr20_+_46213553 | 0.38 |
ENSDART00000100532
|
stx7l
|
syntaxin 7-like |
| chr25_+_31323978 | 0.38 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr12_-_2800809 | 0.37 |
ENSDART00000152682
ENSDART00000083784 |
ubtd1b
|
ubiquitin domain containing 1b |
| chr18_-_46280578 | 0.37 |
ENSDART00000131724
|
pld3
|
phospholipase D family, member 3 |
| chr18_-_7539166 | 0.37 |
ENSDART00000133541
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr19_+_19762183 | 0.37 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr17_+_6563307 | 0.37 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr14_+_23709134 | 0.37 |
ENSDART00000191162
ENSDART00000179754 ENSDART00000054266 |
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr8_+_17168114 | 0.37 |
ENSDART00000183901
|
cenph
|
centromere protein H |
| chr1_-_8140763 | 0.36 |
ENSDART00000091508
|
FAM83G
|
si:dkeyp-9d4.4 |
| chr19_+_19772765 | 0.36 |
ENSDART00000182028
ENSDART00000161019 |
hoxa3a
|
homeobox A3a |
| chr21_+_7298687 | 0.36 |
ENSDART00000187746
|
CU929160.1
|
|
| chr14_+_39156 | 0.36 |
ENSDART00000082184
|
tmem107
|
transmembrane protein 107 |
| chr1_+_57041549 | 0.36 |
ENSDART00000152198
|
si:ch211-1f22.16
|
si:ch211-1f22.16 |
| chr2_+_24507597 | 0.36 |
ENSDART00000133109
|
rps28
|
ribosomal protein S28 |
| chr5_-_54714789 | 0.36 |
ENSDART00000063357
|
ccnb1
|
cyclin B1 |
| chr1_-_18811517 | 0.36 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr8_-_20245892 | 0.36 |
ENSDART00000136911
|
acer1
|
alkaline ceramidase 1 |
| chr25_+_258883 | 0.36 |
ENSDART00000155256
|
zgc:92481
|
zgc:92481 |
| chr3_+_23710839 | 0.36 |
ENSDART00000151584
|
hoxb4a
|
homeobox B4a |
| chr14_+_94946 | 0.36 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr19_+_48176745 | 0.36 |
ENSDART00000164963
|
prdm1b
|
PR domain containing 1b, with ZNF domain |
| chr6_+_153146 | 0.35 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr18_-_7539469 | 0.35 |
ENSDART00000101296
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr4_+_5341592 | 0.35 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr4_-_42242844 | 0.35 |
ENSDART00000163476
|
si:ch211-129p6.2
|
si:ch211-129p6.2 |
| chr3_-_34098731 | 0.35 |
ENSDART00000150999
|
ighv5-3
|
immunoglobulin heavy variable 5-3 |
| chr20_+_32406011 | 0.34 |
ENSDART00000018640
ENSDART00000137910 |
snx3
|
sorting nexin 3 |
| chr2_-_10386738 | 0.34 |
ENSDART00000016369
|
wls
|
wntless Wnt ligand secretion mediator |
| chr23_+_45025909 | 0.34 |
ENSDART00000188105
|
hmgb2b
|
high mobility group box 2b |
| chr20_+_192170 | 0.34 |
ENSDART00000189675
|
cx28.8
|
connexin 28.8 |
| chr3_-_8765165 | 0.33 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr14_+_22076596 | 0.33 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr14_+_41318881 | 0.33 |
ENSDART00000192137
|
xkrx
|
XK, Kell blood group complex subunit-related, X-linked |
| chr6_+_12968101 | 0.33 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr4_-_13614797 | 0.33 |
ENSDART00000138366
ENSDART00000165212 |
irf5
|
interferon regulatory factor 5 |
| chr19_-_1855368 | 0.33 |
ENSDART00000029646
|
rplp1
|
ribosomal protein, large, P1 |
| chr11_-_42472941 | 0.33 |
ENSDART00000166624
|
arf4b
|
ADP-ribosylation factor 4b |
| chr17_+_30843881 | 0.33 |
ENSDART00000149600
ENSDART00000148547 |
tpp1
|
tripeptidyl peptidase I |
| chr3_+_61391636 | 0.33 |
ENSDART00000126417
|
bri3
|
brain protein I3 |
| chr9_+_55857193 | 0.33 |
ENSDART00000160980
|
sept10
|
septin 10 |
| chr14_+_8456870 | 0.33 |
ENSDART00000007738
|
tmco6
|
transmembrane and coiled-coil domains 6 |
| chr17_+_32622933 | 0.33 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr19_+_10493607 | 0.32 |
ENSDART00000003031
|
si:ch211-171h4.6
|
si:ch211-171h4.6 |
| chr15_+_6652396 | 0.32 |
ENSDART00000192813
ENSDART00000157678 |
nop53
|
NOP53 ribosome biogenesis factor |
| chr4_-_68853063 | 0.32 |
ENSDART00000162386
|
si:dkey-264f17.1
|
si:dkey-264f17.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfe3a+tfe3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.3 | 1.3 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.2 | 0.9 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.2 | 1.1 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.2 | 1.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 0.5 | GO:0002544 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.2 | 0.9 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.6 | GO:0070199 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 1.4 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.6 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.1 | 0.4 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 0.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.3 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.3 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.7 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.3 | GO:0019626 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.1 | 0.3 | GO:0051503 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.2 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.1 | 0.2 | GO:0032677 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.5 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.3 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.6 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.2 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.3 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.3 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.2 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.1 | 0.6 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.3 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.1 | 0.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 1.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 1.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.9 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.7 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 1.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.8 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 1.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 0.3 | GO:0021561 | facial nerve development(GO:0021561) |
| 0.0 | 0.1 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.4 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.3 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 1.2 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.8 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 1.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.5 | GO:0090224 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 | 0.4 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.0 | 0.2 | GO:0055016 | hypochord development(GO:0055016) |
| 0.0 | 0.2 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.6 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.5 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.5 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.4 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.0 | 0.1 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:1903332 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.0 | 0.4 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.3 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0070914 | nucleotide-excision repair, DNA gap filling(GO:0006297) UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0098773 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.0 | 0.2 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.5 | GO:1903051 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) negative regulation of proteasomal protein catabolic process(GO:1901799) negative regulation of proteolysis involved in cellular protein catabolic process(GO:1903051) |
| 0.0 | 0.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 1.7 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.8 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.4 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.7 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.1 | GO:0009092 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.0 | 0.3 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.3 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.0 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.0 | 0.9 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.0 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.5 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.6 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.1 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 0.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.3 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.0 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.2 | 0.7 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 0.8 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.6 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.5 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 1.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.6 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.2 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.1 | 0.2 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.1 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.1 | GO:0072380 | TRC complex(GO:0072380) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 1.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.5 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.6 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.9 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.5 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.4 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.0 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.0 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 0.7 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 0.8 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 0.7 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 0.6 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.2 | 0.5 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.4 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.7 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.5 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 0.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.7 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 1.0 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.4 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.4 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.2 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.1 | 0.3 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.3 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.2 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.3 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 2.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.2 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.8 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.6 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 1.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0030250 | calcium sensitive guanylate cyclase activator activity(GO:0008048) cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.1 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.8 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 3.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.5 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.0 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.1 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0031491 | nucleosome binding(GO:0031491) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 0.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.9 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.9 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.4 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 3.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.8 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.0 | 0.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |