Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
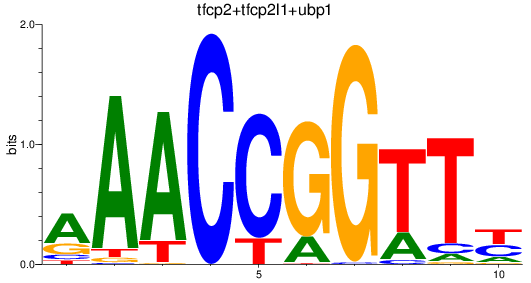
Results for tfcp2+tfcp2l1+ubp1
Z-value: 1.69
Transcription factors associated with tfcp2+tfcp2l1+ubp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ubp1
|
ENSDARG00000018000 | upstream binding protein 1 |
|
tfcp2l1
|
ENSDARG00000029497 | transcription factor CP2-like 1 |
|
tfcp2
|
ENSDARG00000060306 | transcription factor CP2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfcp2l1 | dr11_v1_chr9_+_38292947_38292947 | -0.53 | 3.6e-01 | Click! |
| ubp1 | dr11_v1_chr19_+_42806812_42806814 | -0.17 | 7.9e-01 | Click! |
| tfcp2 | dr11_v1_chr23_-_33679579_33679579 | -0.07 | 9.1e-01 | Click! |
Activity profile of tfcp2+tfcp2l1+ubp1 motif
Sorted Z-values of tfcp2+tfcp2l1+ubp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_51180938 | 1.35 |
ENSDART00000169022
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr10_-_15048781 | 1.31 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr5_+_58665648 | 1.18 |
ENSDART00000167481
|
zgc:194948
|
zgc:194948 |
| chr4_+_76671012 | 1.15 |
ENSDART00000005585
|
ms4a17a.2
|
membrane-spanning 4-domains, subfamily A, member 17a.2 |
| chr5_+_15203421 | 1.05 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr21_+_25688388 | 1.02 |
ENSDART00000125709
|
bicdl2
|
bicaudal-D-related protein 2 |
| chr25_+_10416583 | 1.01 |
ENSDART00000073907
|
ehf
|
ets homologous factor |
| chr3_-_27647845 | 0.94 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr12_-_46145635 | 0.93 |
ENSDART00000074682
|
zgc:153932
|
zgc:153932 |
| chr7_+_73670137 | 0.86 |
ENSDART00000050357
|
btr12
|
bloodthirsty-related gene family, member 12 |
| chr21_-_32436679 | 0.85 |
ENSDART00000076974
|
gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr12_+_38878830 | 0.84 |
ENSDART00000156926
|
si:ch211-39f2.3
|
si:ch211-39f2.3 |
| chr17_-_2039511 | 0.75 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr10_+_39084354 | 0.75 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr9_-_45602978 | 0.73 |
ENSDART00000139019
ENSDART00000085763 |
agr1
|
anterior gradient 1 |
| chr12_+_38929663 | 0.72 |
ENSDART00000156334
|
si:dkey-239b22.1
|
si:dkey-239b22.1 |
| chr15_-_5901514 | 0.72 |
ENSDART00000155252
|
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr4_-_77979432 | 0.71 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr23_+_36115541 | 0.71 |
ENSDART00000130090
|
hoxc6a
|
homeobox C6a |
| chr12_-_46252062 | 0.71 |
ENSDART00000153223
|
si:ch211-226h7.5
|
si:ch211-226h7.5 |
| chr24_+_38671054 | 0.67 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr18_+_47313715 | 0.67 |
ENSDART00000138806
|
barx2
|
BARX homeobox 2 |
| chr3_+_5083407 | 0.67 |
ENSDART00000146883
|
si:ch73-338o16.4
|
si:ch73-338o16.4 |
| chr12_+_22657925 | 0.66 |
ENSDART00000153048
|
si:dkey-219e21.4
|
si:dkey-219e21.4 |
| chr22_-_6404807 | 0.66 |
ENSDART00000149399
ENSDART00000063420 |
si:rp71-1i20.2
|
si:rp71-1i20.2 |
| chr3_-_26204867 | 0.66 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr14_+_17376940 | 0.64 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr15_+_46329149 | 0.64 |
ENSDART00000128404
|
si:ch1073-340i21.3
|
si:ch1073-340i21.3 |
| chr7_+_19835569 | 0.64 |
ENSDART00000149812
|
ovol1a
|
ovo-like zinc finger 1a |
| chr8_+_52479026 | 0.62 |
ENSDART00000162885
|
FO704661.1
|
Danio rerio gamma-glutamyl hydrolase (LOC553228), mRNA. |
| chr16_-_42004544 | 0.62 |
ENSDART00000034544
|
caspa
|
caspase a |
| chr18_+_47313899 | 0.61 |
ENSDART00000192389
ENSDART00000189592 ENSDART00000184281 |
barx2
|
BARX homeobox 2 |
| chr12_-_7607114 | 0.57 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr6_-_24103666 | 0.57 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr7_-_56606752 | 0.57 |
ENSDART00000138714
|
sult5a1
|
sulfotransferase family 5A, member 1 |
| chr5_-_37116265 | 0.57 |
ENSDART00000057613
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr15_+_22014029 | 0.56 |
ENSDART00000079504
|
ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr12_-_28794957 | 0.56 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr20_+_26916639 | 0.56 |
ENSDART00000077787
|
serpinb1l2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 2 |
| chr5_+_30741730 | 0.55 |
ENSDART00000098246
ENSDART00000186992 ENSDART00000182533 |
ftr83
|
finTRIM family, member 83 |
| chr12_-_16990896 | 0.55 |
ENSDART00000152402
|
ifit12
|
interferon-induced protein with tetratricopeptide repeats 12 |
| chr19_+_40856534 | 0.54 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr7_+_20503344 | 0.54 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr12_+_27156943 | 0.54 |
ENSDART00000153030
ENSDART00000001737 |
skap1
|
src kinase associated phosphoprotein 1 |
| chr8_-_28349859 | 0.53 |
ENSDART00000062671
|
tuba8l
|
tubulin, alpha 8 like |
| chr19_+_40856807 | 0.53 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr20_-_39367895 | 0.53 |
ENSDART00000136476
ENSDART00000021788 ENSDART00000180784 |
pbk
|
PDZ binding kinase |
| chr18_-_40773413 | 0.53 |
ENSDART00000133797
|
vaspb
|
vasodilator stimulated phosphoprotein b |
| chr17_-_37395460 | 0.52 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr3_+_6469754 | 0.52 |
ENSDART00000185809
|
NUP85 (1 of many)
|
nucleoporin 85 |
| chr13_-_25774183 | 0.52 |
ENSDART00000046981
|
pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr15_-_452347 | 0.52 |
ENSDART00000115233
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr9_-_443451 | 0.52 |
ENSDART00000165642
|
si:dkey-11f4.14
|
si:dkey-11f4.14 |
| chr12_+_46634736 | 0.52 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr12_-_46112892 | 0.51 |
ENSDART00000187128
ENSDART00000114268 |
zgc:153932
|
zgc:153932 |
| chr21_+_25625026 | 0.50 |
ENSDART00000134678
|
ovol1b
|
ovo-like zinc finger 1b |
| chr15_-_34892664 | 0.50 |
ENSDART00000153787
ENSDART00000099721 |
rnf183
|
ring finger protein 183 |
| chr2_-_43151358 | 0.49 |
ENSDART00000061170
ENSDART00000056161 |
ftr15
|
finTRIM family, member 15 |
| chr9_-_11550711 | 0.48 |
ENSDART00000093343
|
fev
|
FEV (ETS oncogene family) |
| chr5_+_26204561 | 0.48 |
ENSDART00000137178
|
marveld2b
|
MARVEL domain containing 2b |
| chr8_-_52715911 | 0.47 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr20_+_16881883 | 0.47 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr13_+_18533005 | 0.46 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr22_+_22021936 | 0.46 |
ENSDART00000149586
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr4_+_78058635 | 0.45 |
ENSDART00000162631
ENSDART00000191619 |
CABZ01075274.1
|
|
| chr22_-_17631675 | 0.45 |
ENSDART00000132565
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr23_-_3511630 | 0.44 |
ENSDART00000019667
|
rnf114
|
ring finger protein 114 |
| chr23_+_7471072 | 0.44 |
ENSDART00000135551
|
si:ch211-200e2.1
|
si:ch211-200e2.1 |
| chr14_-_25956804 | 0.44 |
ENSDART00000135627
ENSDART00000146022 ENSDART00000039660 |
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr10_+_36662640 | 0.43 |
ENSDART00000063359
|
ucp2
|
uncoupling protein 2 |
| chr8_-_20914829 | 0.43 |
ENSDART00000025356
|
haus5
|
HAUS augmin-like complex, subunit 5 |
| chr22_-_17499513 | 0.43 |
ENSDART00000105460
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr18_+_27571448 | 0.42 |
ENSDART00000147886
|
cd82b
|
CD82 molecule b |
| chr19_-_42588510 | 0.40 |
ENSDART00000102583
|
sytl1
|
synaptotagmin-like 1 |
| chr7_-_24193051 | 0.39 |
ENSDART00000169737
ENSDART00000171085 |
si:ch211-216p19.6
|
si:ch211-216p19.6 |
| chr3_-_53092509 | 0.39 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr22_+_7497319 | 0.38 |
ENSDART00000034564
|
CELA1 (1 of many)
|
zgc:92511 |
| chr3_-_34113838 | 0.38 |
ENSDART00000151216
|
ighv4-2
|
immunoglobulin heavy variable 4-2 |
| chr5_+_21891305 | 0.38 |
ENSDART00000136788
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr5_+_38631687 | 0.38 |
ENSDART00000147280
|
si:ch211-271e10.6
|
si:ch211-271e10.6 |
| chr8_+_52377516 | 0.37 |
ENSDART00000115398
|
arid5a
|
AT rich interactive domain 5A (MRF1-like) |
| chr15_-_28904371 | 0.36 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr19_-_47570672 | 0.35 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr2_-_47620806 | 0.35 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr7_+_57089354 | 0.35 |
ENSDART00000140702
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr17_+_51743908 | 0.34 |
ENSDART00000149039
ENSDART00000148869 |
odc1
|
ornithine decarboxylase 1 |
| chr5_-_64883082 | 0.34 |
ENSDART00000064983
ENSDART00000139066 |
krt1-c5
|
keratin, type 1, gene c5 |
| chr19_+_31576849 | 0.34 |
ENSDART00000134107
ENSDART00000088401 |
acot13
|
acyl-CoA thioesterase 13 |
| chr5_+_36599107 | 0.33 |
ENSDART00000097677
ENSDART00000138147 |
micu2
|
mitochondrial calcium uptake 2 |
| chr16_+_23961276 | 0.33 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr24_+_20960216 | 0.33 |
ENSDART00000133008
|
si:ch211-161h7.8
|
si:ch211-161h7.8 |
| chr5_+_38623814 | 0.33 |
ENSDART00000146046
|
si:ch211-271e10.3
|
si:ch211-271e10.3 |
| chr10_-_36793412 | 0.33 |
ENSDART00000185966
|
dhrs13a.2
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 2 |
| chr13_+_669177 | 0.32 |
ENSDART00000190085
|
CU462915.1
|
|
| chr8_+_36142734 | 0.32 |
ENSDART00000159361
ENSDART00000161194 |
mhc2b
|
major histocompatibility complex class II integral membrane beta chain gene |
| chr6_-_54486410 | 0.32 |
ENSDART00000064679
|
CR847944.1
|
|
| chr15_+_12429206 | 0.32 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr10_-_1523253 | 0.32 |
ENSDART00000179510
ENSDART00000176548 ENSDART00000180368 ENSDART00000185270 |
WDR70
|
WD repeat domain 70 |
| chr21_-_42831033 | 0.32 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr5_+_20319519 | 0.31 |
ENSDART00000004217
|
coro1ca
|
coronin, actin binding protein, 1Ca |
| chr14_+_94603 | 0.31 |
ENSDART00000162480
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr11_-_36350876 | 0.31 |
ENSDART00000146495
ENSDART00000020655 |
psma5
|
proteasome subunit alpha 5 |
| chr8_-_3312384 | 0.31 |
ENSDART00000035965
|
fut9b
|
fucosyltransferase 9b |
| chr7_+_57725708 | 0.30 |
ENSDART00000056466
ENSDART00000142259 ENSDART00000166198 |
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr3_+_41585298 | 0.29 |
ENSDART00000182370
|
card11
|
caspase recruitment domain family, member 11 |
| chr22_-_3299355 | 0.29 |
ENSDART00000190993
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr18_-_8877077 | 0.28 |
ENSDART00000137266
|
si:dkey-95h12.2
|
si:dkey-95h12.2 |
| chr11_-_309420 | 0.28 |
ENSDART00000173185
|
poc1a
|
POC1 centriolar protein A |
| chr22_+_9806867 | 0.28 |
ENSDART00000137182
ENSDART00000106103 |
si:ch211-236g6.1
|
si:ch211-236g6.1 |
| chr9_-_3400727 | 0.28 |
ENSDART00000183979
ENSDART00000111386 |
dlx2a
|
distal-less homeobox 2a |
| chr20_-_25631256 | 0.28 |
ENSDART00000048164
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr19_-_2861444 | 0.27 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr7_+_24205034 | 0.27 |
ENSDART00000018580
|
nedd8
|
neural precursor cell expressed, developmentally down-regulated 8 |
| chr3_-_23512285 | 0.27 |
ENSDART00000159151
|
BX682558.1
|
|
| chr4_+_6032640 | 0.27 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr1_-_21321482 | 0.26 |
ENSDART00000054440
|
tmem144a
|
transmembrane protein 144a |
| chr7_-_21928826 | 0.26 |
ENSDART00000088043
|
si:dkey-85k7.11
|
si:dkey-85k7.11 |
| chr20_+_53181017 | 0.26 |
ENSDART00000189692
ENSDART00000177109 |
FIG4
|
FIG4 phosphoinositide 5-phosphatase |
| chr16_+_29586468 | 0.25 |
ENSDART00000148926
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr22_-_34979139 | 0.25 |
ENSDART00000116455
ENSDART00000133537 |
arhgap19
|
Rho GTPase activating protein 19 |
| chr4_-_77636185 | 0.25 |
ENSDART00000152915
ENSDART00000131964 |
si:dkey-61p9.9
|
si:dkey-61p9.9 |
| chr7_+_54259657 | 0.25 |
ENSDART00000170174
|
pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr8_+_25342896 | 0.25 |
ENSDART00000129032
|
CR847543.1
|
|
| chr14_+_9421510 | 0.24 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr8_+_27555314 | 0.24 |
ENSDART00000135568
ENSDART00000016696 |
rhocb
|
ras homolog family member Cb |
| chr2_-_42035250 | 0.24 |
ENSDART00000056460
ENSDART00000140788 |
gbp1
|
guanylate binding protein 1 |
| chr3_-_29977495 | 0.24 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr6_+_13506841 | 0.24 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr5_-_30418636 | 0.24 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr10_+_44692272 | 0.23 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr5_-_54712159 | 0.23 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr2_-_9527129 | 0.23 |
ENSDART00000157422
ENSDART00000004398 |
cope
|
coatomer protein complex, subunit epsilon |
| chr6_+_23810529 | 0.23 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr7_+_57088920 | 0.23 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr15_+_41919484 | 0.23 |
ENSDART00000099821
ENSDART00000146246 |
nlrp16
|
NACHT, LRR and PYD domains-containing protein 16 |
| chr24_-_26479841 | 0.23 |
ENSDART00000079984
|
rpl22l1
|
ribosomal protein L22-like 1 |
| chr13_+_33268657 | 0.23 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr1_+_51475094 | 0.22 |
ENSDART00000146352
|
meis1a
|
Meis homeobox 1 a |
| chr9_+_21260314 | 0.22 |
ENSDART00000145943
|
ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr7_+_25221757 | 0.22 |
ENSDART00000173551
|
exoc6b
|
exocyst complex component 6B |
| chr2_-_7711522 | 0.22 |
ENSDART00000163175
|
dcun1d1
|
DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) |
| chr8_-_46457233 | 0.22 |
ENSDART00000113214
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr22_+_19552987 | 0.22 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr16_+_27224369 | 0.22 |
ENSDART00000136980
|
sec61b
|
Sec61 translocon beta subunit |
| chr18_+_5308392 | 0.21 |
ENSDART00000179072
|
DUT
|
deoxyuridine triphosphatase |
| chr6_+_41452979 | 0.21 |
ENSDART00000007353
|
wdr82
|
WD repeat domain 82 |
| chr13_-_2112450 | 0.21 |
ENSDART00000189343
|
fam83b
|
family with sequence similarity 83, member B |
| chr6_-_27108844 | 0.21 |
ENSDART00000073883
|
dtymk
|
deoxythymidylate kinase (thymidylate kinase) |
| chr11_+_3246059 | 0.21 |
ENSDART00000161529
|
timeless
|
timeless circadian clock |
| chr16_+_29509133 | 0.20 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr20_-_14012859 | 0.20 |
ENSDART00000152429
|
si:ch211-22i13.2
|
si:ch211-22i13.2 |
| chr10_+_41169996 | 0.20 |
ENSDART00000189737
|
anxa4
|
annexin A4 |
| chr1_-_58868306 | 0.20 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr11_-_36350421 | 0.20 |
ENSDART00000141477
|
psma5
|
proteasome subunit alpha 5 |
| chr23_-_18567088 | 0.20 |
ENSDART00000192371
|
sephs2
|
selenophosphate synthetase 2 |
| chr11_-_308838 | 0.20 |
ENSDART00000112538
|
poc1a
|
POC1 centriolar protein A |
| chr9_-_10778615 | 0.20 |
ENSDART00000182577
|
CABZ01053619.1
|
|
| chr15_-_4596623 | 0.20 |
ENSDART00000132227
|
eif4h
|
eukaryotic translation initiation factor 4h |
| chr18_-_7448047 | 0.19 |
ENSDART00000193213
ENSDART00000131940 ENSDART00000186944 ENSDART00000052803 |
si:dkey-30c15.10
|
si:dkey-30c15.10 |
| chr14_+_7048930 | 0.19 |
ENSDART00000109138
|
hbegfa
|
heparin-binding EGF-like growth factor a |
| chr1_-_56556326 | 0.19 |
ENSDART00000188760
|
CABZ01059408.1
|
|
| chr20_-_40729364 | 0.19 |
ENSDART00000101014
|
cx32.2
|
connexin 32.2 |
| chr4_+_47751483 | 0.19 |
ENSDART00000160043
|
si:ch211-196f19.1
|
si:ch211-196f19.1 |
| chr20_+_7584211 | 0.18 |
ENSDART00000132481
ENSDART00000127975 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr12_+_30234209 | 0.18 |
ENSDART00000102081
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr7_-_54430505 | 0.18 |
ENSDART00000167905
|
ano1
|
anoctamin 1, calcium activated chloride channel |
| chr15_-_34668485 | 0.18 |
ENSDART00000186605
|
bag6
|
BCL2 associated athanogene 6 |
| chr1_-_54063520 | 0.18 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr22_-_3299100 | 0.18 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr23_+_36144487 | 0.18 |
ENSDART00000082473
|
hoxc3a
|
homeobox C3a |
| chr2_-_36600943 | 0.17 |
ENSDART00000158597
|
tradv36.2.12
|
T-cell receptor alpha/delta variable 36.2.12 |
| chr14_+_45350008 | 0.17 |
ENSDART00000025749
|
ttc9c
|
tetratricopeptide repeat domain 9C |
| chr11_+_41858807 | 0.17 |
ENSDART00000161605
|
iffo2b
|
intermediate filament family orphan 2b |
| chr7_-_34256374 | 0.17 |
ENSDART00000075176
|
dis3l
|
DIS3 like exosome 3'-5' exoribonuclease |
| chr22_-_19504690 | 0.17 |
ENSDART00000166296
|
si:dkey-78l4.11
|
si:dkey-78l4.11 |
| chr16_+_45739193 | 0.17 |
ENSDART00000184852
ENSDART00000156851 ENSDART00000154704 |
paqr6
|
progestin and adipoQ receptor family member VI |
| chr2_+_53720028 | 0.16 |
ENSDART00000170799
|
ctnnbl1
|
catenin, beta like 1 |
| chr14_+_41409697 | 0.16 |
ENSDART00000173335
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr13_+_24671481 | 0.16 |
ENSDART00000001678
|
adam8a
|
ADAM metallopeptidase domain 8a |
| chr8_-_31716302 | 0.16 |
ENSDART00000061832
|
si:dkey-46a10.3
|
si:dkey-46a10.3 |
| chr23_-_26252103 | 0.16 |
ENSDART00000160873
|
magi3a
|
membrane associated guanylate kinase, WW and PDZ domain containing 3a |
| chr8_-_418997 | 0.16 |
ENSDART00000166683
|
pggt1b
|
protein geranylgeranyltransferase type I, beta subunit |
| chr4_+_64034834 | 0.16 |
ENSDART00000160654
|
si:dkey-179k24.5
|
si:dkey-179k24.5 |
| chr13_-_36535128 | 0.15 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr6_+_149405 | 0.15 |
ENSDART00000161154
|
fdx1l
|
ferredoxin 1-like |
| chr11_-_21303946 | 0.15 |
ENSDART00000185786
|
RASSF5
|
si:dkey-85p17.3 |
| chr1_-_54191626 | 0.15 |
ENSDART00000062941
|
nthl1
|
nth-like DNA glycosylase 1 |
| chr24_-_30275204 | 0.15 |
ENSDART00000164187
|
snx7
|
sorting nexin 7 |
| chr4_+_43700319 | 0.15 |
ENSDART00000141967
|
si:ch211-226o13.1
|
si:ch211-226o13.1 |
| chr6_+_72040 | 0.15 |
ENSDART00000122957
|
crygm4
|
crystallin, gamma M4 |
| chr23_-_19153378 | 0.15 |
ENSDART00000019045
ENSDART00000183681 |
ebp
|
emopamil binding protein (sterol isomerase) |
| chr6_-_49526510 | 0.14 |
ENSDART00000128025
|
rps26l
|
ribosomal protein S26, like |
| chr1_-_11104805 | 0.14 |
ENSDART00000147648
|
knl1
|
kinetochore scaffold 1 |
| chr7_+_19483277 | 0.14 |
ENSDART00000173750
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr15_-_41486793 | 0.14 |
ENSDART00000138525
|
si:ch211-187g4.1
|
si:ch211-187g4.1 |
| chr2_-_58201173 | 0.14 |
ENSDART00000166282
|
pnp5b
|
purine nucleoside phosphorylase 5b |
| chr8_-_49725430 | 0.14 |
ENSDART00000135675
|
gkap1
|
G kinase anchoring protein 1 |
| chr15_+_13984879 | 0.14 |
ENSDART00000159438
|
zgc:162730
|
zgc:162730 |
| chr8_-_29658171 | 0.14 |
ENSDART00000147755
|
mpeg1.3
|
macrophage expressed 1, tandem duplicate 3 |
| chr10_-_7988396 | 0.13 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfcp2+tfcp2l1+ubp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.3 | 1.0 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.2 | 0.5 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.2 | 0.5 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 0.3 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.1 | 0.3 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.6 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.1 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 0.2 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.1 | 0.9 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.3 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.3 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.3 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.3 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.0 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.2 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.0 | 0.4 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.4 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.3 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.2 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.7 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.0 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.0 | 3.0 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.0 | 0.4 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.1 | GO:0003151 | outflow tract morphogenesis(GO:0003151) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.6 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.5 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.3 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0008210 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.0 | 0.1 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.2 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.4 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 0.1 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.0 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.0 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 1.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.2 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.2 | 3.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.4 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.3 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.2 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.3 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 1.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.4 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 0.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.3 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.1 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.7 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.8 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |