Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
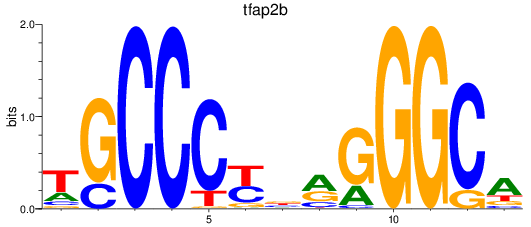
Results for tfap2b
Z-value: 1.26
Transcription factors associated with tfap2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfap2b
|
ENSDARG00000012667 | transcription factor AP-2 beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfap2b | dr11_v1_chr20_-_47704973_47704973 | 0.38 | 5.2e-01 | Click! |
Activity profile of tfap2b motif
Sorted Z-values of tfap2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_26612401 | 0.71 |
ENSDART00000145571
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr21_+_26748141 | 0.65 |
ENSDART00000169025
|
pcxa
|
pyruvate carboxylase a |
| chr3_+_15271943 | 0.57 |
ENSDART00000141752
|
asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr15_-_32131 | 0.54 |
ENSDART00000099074
ENSDART00000164323 |
CYP2C9
|
si:zfos-411a11.2 |
| chr7_+_34794829 | 0.52 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr4_-_77551860 | 0.46 |
ENSDART00000188176
|
AL935186.6
|
|
| chr15_-_20710 | 0.43 |
ENSDART00000161218
|
cyp2y3
|
cytochrome P450, family 2, subfamily Y, polypeptide 3 |
| chr4_-_77563411 | 0.42 |
ENSDART00000186841
|
AL935186.8
|
|
| chr21_-_27362938 | 0.42 |
ENSDART00000131297
|
rin1a
|
Ras and Rab interactor 1a |
| chr24_-_38374744 | 0.42 |
ENSDART00000007208
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr18_+_24919614 | 0.37 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr12_-_8958353 | 0.36 |
ENSDART00000041728
|
cyp26a1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr2_-_42415902 | 0.36 |
ENSDART00000142489
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr22_-_17953529 | 0.35 |
ENSDART00000135716
|
ncanb
|
neurocan b |
| chr21_-_34926619 | 0.34 |
ENSDART00000065337
ENSDART00000192740 |
kif20a
|
kinesin family member 20A |
| chr7_+_529522 | 0.34 |
ENSDART00000190811
|
nrxn2b
|
neurexin 2b |
| chr10_-_43964028 | 0.32 |
ENSDART00000009134
ENSDART00000133450 |
sept5b
|
septin 5b |
| chr13_+_16521898 | 0.32 |
ENSDART00000122557
|
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr12_+_35119762 | 0.32 |
ENSDART00000085774
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr23_-_1017605 | 0.30 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr21_+_21279159 | 0.30 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr16_-_20435475 | 0.30 |
ENSDART00000139776
|
chn2
|
chimerin 2 |
| chr6_+_38381957 | 0.26 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr23_-_1017428 | 0.26 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr18_+_18104235 | 0.25 |
ENSDART00000145342
|
cbln1
|
cerebellin 1 precursor |
| chr14_+_10450216 | 0.25 |
ENSDART00000134814
|
cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr8_+_29963024 | 0.24 |
ENSDART00000149372
|
ptch1
|
patched 1 |
| chr20_-_54014373 | 0.24 |
ENSDART00000152934
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr12_+_24562667 | 0.24 |
ENSDART00000056256
|
nrxn1a
|
neurexin 1a |
| chr25_+_22017182 | 0.23 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr25_+_8063455 | 0.23 |
ENSDART00000073919
|
kcnc1b
|
potassium voltage-gated channel, Shaw-related subfamily, member 1b |
| chr11_-_33857911 | 0.23 |
ENSDART00000165370
|
nxph2b
|
neurexophilin 2b |
| chr6_+_102506 | 0.22 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr17_-_26868169 | 0.22 |
ENSDART00000157204
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr11_-_35975026 | 0.21 |
ENSDART00000186219
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr23_-_27701361 | 0.21 |
ENSDART00000186688
ENSDART00000183985 |
dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr4_+_58667348 | 0.21 |
ENSDART00000186596
ENSDART00000180673 |
CR388148.2
|
|
| chr1_+_32528097 | 0.21 |
ENSDART00000128317
|
nlgn4a
|
neuroligin 4a |
| chr17_-_19019635 | 0.20 |
ENSDART00000126666
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr2_-_34555945 | 0.20 |
ENSDART00000056671
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr25_+_31264155 | 0.19 |
ENSDART00000012256
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr5_+_45921234 | 0.19 |
ENSDART00000134355
|
ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr17_-_26867725 | 0.18 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr24_+_37723362 | 0.18 |
ENSDART00000136836
|
rab11fip3
|
RAB11 family interacting protein 3 (class II) |
| chr25_+_21180695 | 0.18 |
ENSDART00000012923
|
erc1a
|
ELKS/RAB6-interacting/CAST family member 1a |
| chr22_+_508290 | 0.18 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr2_-_9544161 | 0.18 |
ENSDART00000124425
|
slc25a24l
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24, like |
| chr14_+_11103718 | 0.17 |
ENSDART00000161311
|
nexmifb
|
neurite extension and migration factor b |
| chr7_+_1442059 | 0.17 |
ENSDART00000173391
|
si:cabz01090193.1
|
si:cabz01090193.1 |
| chr3_+_16455749 | 0.16 |
ENSDART00000090767
|
kcnj12b
|
potassium inwardly-rectifying channel, subfamily J, member 12b |
| chr25_+_16945348 | 0.15 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr5_-_15948833 | 0.14 |
ENSDART00000051649
ENSDART00000124467 |
xbp1
|
X-box binding protein 1 |
| chr1_-_55248496 | 0.13 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr13_+_36585399 | 0.13 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr23_+_39461989 | 0.13 |
ENSDART00000184761
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr2_+_39108339 | 0.11 |
ENSDART00000085675
|
clstn2
|
calsyntenin 2 |
| chr21_+_21205667 | 0.11 |
ENSDART00000058311
|
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr10_-_27197044 | 0.11 |
ENSDART00000137928
|
auts2a
|
autism susceptibility candidate 2a |
| chr12_-_21834611 | 0.11 |
ENSDART00000179553
|
thrab
|
thyroid hormone receptor alpha b |
| chr25_-_32363341 | 0.11 |
ENSDART00000153892
ENSDART00000114385 |
cep152
|
centrosomal protein 152 |
| chr25_+_6471401 | 0.10 |
ENSDART00000132927
|
snx33
|
sorting nexin 33 |
| chr6_+_7322587 | 0.10 |
ENSDART00000065500
|
abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr19_+_41080240 | 0.10 |
ENSDART00000087295
|
ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr12_+_5708400 | 0.10 |
ENSDART00000017191
|
dlx3b
|
distal-less homeobox 3b |
| chr5_+_7393346 | 0.09 |
ENSDART00000164238
|
si:ch73-72b7.1
|
si:ch73-72b7.1 |
| chr25_-_26753196 | 0.09 |
ENSDART00000155698
|
usp3
|
ubiquitin specific peptidase 3 |
| chr20_-_3160687 | 0.08 |
ENSDART00000131697
|
si:ch73-212j7.3
|
si:ch73-212j7.3 |
| chr16_-_21620947 | 0.08 |
ENSDART00000115011
ENSDART00000183125 ENSDART00000188856 ENSDART00000189460 |
ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr22_-_9906790 | 0.08 |
ENSDART00000105936
|
si:dkey-253d23.8
|
si:dkey-253d23.8 |
| chr12_-_27242498 | 0.08 |
ENSDART00000152609
ENSDART00000152170 |
si:dkey-11c5.11
|
si:dkey-11c5.11 |
| chr12_+_30360184 | 0.07 |
ENSDART00000190718
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
| chr25_+_23489979 | 0.07 |
ENSDART00000158200
|
CU024870.1
|
|
| chr18_+_40364732 | 0.07 |
ENSDART00000123661
ENSDART00000130847 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr15_-_45246911 | 0.07 |
ENSDART00000189557
ENSDART00000185291 |
CABZ01072607.1
|
|
| chr6_-_19035749 | 0.07 |
ENSDART00000187714
|
sept9b
|
septin 9b |
| chr16_+_54209504 | 0.06 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr14_-_51039702 | 0.05 |
ENSDART00000178932
|
cltb
|
clathrin, light chain B |
| chr13_-_35497586 | 0.05 |
ENSDART00000148402
|
srd5a2b
|
steroid-5-alpha-reductase, alpha polypeptide 2b |
| chr12_-_32013125 | 0.05 |
ENSDART00000153355
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr7_-_8881514 | 0.05 |
ENSDART00000081620
|
vax2
|
ventral anterior homeobox 2 |
| chr7_-_7420301 | 0.05 |
ENSDART00000102620
|
six7
|
SIX homeobox 7 |
| chr10_-_5135788 | 0.04 |
ENSDART00000108587
ENSDART00000138537 |
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr3_-_43646733 | 0.04 |
ENSDART00000180959
|
axin1
|
axin 1 |
| chr20_-_47188966 | 0.04 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr1_+_58990121 | 0.04 |
ENSDART00000171654
|
CABZ01083166.1
|
|
| chr17_-_27972307 | 0.03 |
ENSDART00000062130
|
ptafr
|
platelet-activating factor receptor |
| chr4_-_63980140 | 0.03 |
ENSDART00000160818
|
znf1091
|
zinc finger protein 1091 |
| chr17_-_29771639 | 0.03 |
ENSDART00000086201
|
ush2a
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr12_+_30360579 | 0.03 |
ENSDART00000152900
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
| chr10_+_6383270 | 0.03 |
ENSDART00000170548
|
zgc:114200
|
zgc:114200 |
| chr1_+_18863060 | 0.03 |
ENSDART00000139241
|
rnf38
|
ring finger protein 38 |
| chr6_+_9867426 | 0.02 |
ENSDART00000151749
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr6_-_59942335 | 0.02 |
ENSDART00000168416
|
fbxl3b
|
F-box and leucine-rich repeat protein 3b |
| chr20_+_51199666 | 0.02 |
ENSDART00000169321
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr25_-_20870900 | 0.02 |
ENSDART00000171288
|
magi2b
|
membrane associated guanylate kinase, WW and PDZ domain containing 2b |
| chr23_+_25868637 | 0.01 |
ENSDART00000103934
|
ttpal
|
tocopherol (alpha) transfer protein-like |
| chr21_+_39292266 | 0.01 |
ENSDART00000144116
|
si:ch211-274p24.2
|
si:ch211-274p24.2 |
| chr5_-_7513082 | 0.01 |
ENSDART00000158913
|
bmpr1ba
|
bone morphogenetic protein receptor, type IBa |
| chr7_+_1812480 | 0.01 |
ENSDART00000173278
|
si:ch73-126o18.1
|
si:ch73-126o18.1 |
| chr9_+_33334501 | 0.00 |
ENSDART00000006867
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr9_+_44721808 | 0.00 |
ENSDART00000190578
|
nckap1
|
NCK-associated protein 1 |
| chr15_-_18213515 | 0.00 |
ENSDART00000101635
|
btr21
|
bloodthirsty-related gene family, member 21 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfap2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.4 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.1 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.0 | 0.2 | GO:2000048 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.2 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.4 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.0 | GO:0030910 | olfactory placode formation(GO:0030910) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043220 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.4 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.4 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |