Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
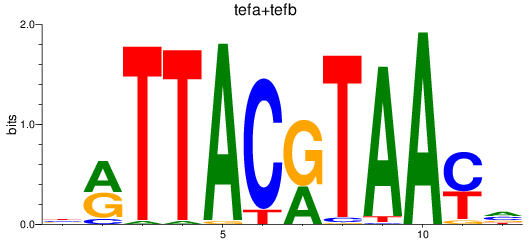
Results for tefa+tefb
Z-value: 1.73
Transcription factors associated with tefa+tefb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tefa
|
ENSDARG00000039117 | TEF transcription factor, PAR bZIP family member a |
|
tefb
|
ENSDARG00000098103 | TEF transcription factor, PAR bZIP family member b |
|
tefa
|
ENSDARG00000109532 | TEF transcription factor, PAR bZIP family member a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tefb | dr11_v1_chr3_-_5067585_5067585 | 0.87 | 5.2e-02 | Click! |
| tefa | dr11_v1_chr12_-_19151708_19151708 | 0.53 | 3.6e-01 | Click! |
Activity profile of tefa+tefb motif
Sorted Z-values of tefa+tefb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_15354095 | 2.70 |
ENSDART00000090397
|
kiaa1549la
|
KIAA1549-like a |
| chr1_+_31112436 | 2.59 |
ENSDART00000075340
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr16_+_39146696 | 2.25 |
ENSDART00000121756
ENSDART00000084381 |
sybu
|
syntabulin (syntaxin-interacting) |
| chr12_-_26383242 | 2.09 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr16_+_34523515 | 2.09 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr17_-_43466317 | 2.05 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr4_-_5302866 | 1.95 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr16_+_46111849 | 1.94 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr3_-_46818001 | 1.94 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr17_-_20979077 | 1.83 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr3_-_28665291 | 1.80 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr11_-_10770053 | 1.80 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr5_-_22001003 | 1.74 |
ENSDART00000134393
ENSDART00000143878 |
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr5_-_21970881 | 1.71 |
ENSDART00000182907
|
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr4_-_5302162 | 1.64 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr7_+_38717624 | 1.63 |
ENSDART00000132522
|
syt13
|
synaptotagmin XIII |
| chr10_-_24371312 | 1.61 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr2_+_24177190 | 1.54 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr20_+_38201644 | 1.38 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr20_-_45661049 | 1.36 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr10_+_33895315 | 1.36 |
ENSDART00000142881
|
fryb
|
furry homolog b (Drosophila) |
| chr9_+_17862858 | 1.36 |
ENSDART00000166566
|
dgkh
|
diacylglycerol kinase, eta |
| chr20_-_40717900 | 1.32 |
ENSDART00000181663
|
cx43
|
connexin 43 |
| chr5_+_1278092 | 1.26 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr14_+_34971554 | 1.26 |
ENSDART00000184271
|
rnf145a
|
ring finger protein 145a |
| chr25_-_11088839 | 1.22 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr7_+_38278860 | 1.13 |
ENSDART00000016265
|
lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr24_-_39772045 | 1.12 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr22_-_11438627 | 1.09 |
ENSDART00000007649
|
mid1ip1b
|
MID1 interacting protein 1b |
| chr9_+_46644633 | 1.06 |
ENSDART00000160285
|
slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr7_-_6604623 | 1.06 |
ENSDART00000172874
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr21_-_32467799 | 1.04 |
ENSDART00000007675
ENSDART00000133099 |
zgc:123105
|
zgc:123105 |
| chr11_+_6819050 | 1.00 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr20_+_1121458 | 1.00 |
ENSDART00000064472
|
pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr1_+_54673846 | 0.99 |
ENSDART00000145018
|
gprc5bb
|
G protein-coupled receptor, class C, group 5, member Bb |
| chr21_-_42202792 | 0.99 |
ENSDART00000124708
|
gabra6b
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6b |
| chr5_+_15350954 | 0.99 |
ENSDART00000140990
ENSDART00000137287 ENSDART00000061653 |
pebp1
|
phosphatidylethanolamine binding protein 1 |
| chr8_+_8712446 | 0.97 |
ENSDART00000158674
|
elk1
|
ELK1, member of ETS oncogene family |
| chr18_-_39288894 | 0.92 |
ENSDART00000186216
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr7_+_17716601 | 0.91 |
ENSDART00000173792
ENSDART00000080825 |
rtn3
|
reticulon 3 |
| chr16_-_45069882 | 0.86 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr14_+_36628131 | 0.85 |
ENSDART00000188625
ENSDART00000125345 |
TENM3
|
si:dkey-237h12.3 |
| chr3_-_21348478 | 0.83 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr5_-_21030934 | 0.83 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr21_-_32467099 | 0.82 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr19_+_7549854 | 0.79 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr25_-_13490744 | 0.77 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr7_-_32833153 | 0.77 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr20_-_1151265 | 0.72 |
ENSDART00000012376
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr18_-_1228688 | 0.72 |
ENSDART00000064403
|
nptnb
|
neuroplastin b |
| chr8_-_29851706 | 0.67 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr20_+_3934516 | 0.67 |
ENSDART00000165732
|
clec11a
|
C-type lectin domain containing 11A |
| chr20_-_44576949 | 0.65 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr9_+_33788626 | 0.64 |
ENSDART00000088469
|
kdm6a
|
lysine (K)-specific demethylase 6A |
| chr24_+_18948665 | 0.62 |
ENSDART00000106186
|
prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr10_-_26729930 | 0.60 |
ENSDART00000145532
|
fgf13b
|
fibroblast growth factor 13b |
| chr17_+_15845765 | 0.59 |
ENSDART00000130881
ENSDART00000074936 |
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr20_+_23083800 | 0.59 |
ENSDART00000132093
|
usp46
|
ubiquitin specific peptidase 46 |
| chr6_-_29612269 | 0.52 |
ENSDART00000104293
|
pex5la
|
peroxisomal biogenesis factor 5-like a |
| chr9_-_13871935 | 0.52 |
ENSDART00000146597
|
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr7_+_52761841 | 0.47 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr15_-_20939579 | 0.45 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr12_-_20584413 | 0.45 |
ENSDART00000170923
|
FP885542.2
|
|
| chr9_-_9225980 | 0.45 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr14_+_23717165 | 0.44 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr22_-_18491813 | 0.41 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr21_+_43178831 | 0.41 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr7_-_38792543 | 0.41 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr17_-_8268406 | 0.40 |
ENSDART00000149873
ENSDART00000064668 ENSDART00000148403 |
ahi1
|
Abelson helper integration site 1 |
| chr7_-_40959667 | 0.40 |
ENSDART00000084070
|
rbm33a
|
RNA binding motif protein 33a |
| chr11_-_7078392 | 0.39 |
ENSDART00000112156
ENSDART00000188556 |
si:ch211-253b8.5
|
si:ch211-253b8.5 |
| chr5_+_69808763 | 0.38 |
ENSDART00000143482
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr21_-_39566854 | 0.36 |
ENSDART00000020174
|
dynll2b
|
dynein, light chain, LC8-type 2b |
| chr9_+_33788389 | 0.36 |
ENSDART00000144623
|
kdm6a
|
lysine (K)-specific demethylase 6A |
| chr12_+_36416173 | 0.35 |
ENSDART00000190278
|
unk
|
unkempt family zinc finger |
| chr19_-_45650994 | 0.35 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr5_-_65153731 | 0.34 |
ENSDART00000171024
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr15_+_37559570 | 0.34 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr1_+_19764995 | 0.33 |
ENSDART00000138276
|
si:ch211-42i9.8
|
si:ch211-42i9.8 |
| chr23_-_21763598 | 0.33 |
ENSDART00000145408
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr20_-_44496245 | 0.33 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr23_+_33963619 | 0.33 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr16_-_35427060 | 0.31 |
ENSDART00000172294
|
ctps1b
|
CTP synthase 1b |
| chr25_-_16826219 | 0.30 |
ENSDART00000191299
ENSDART00000188504 |
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr22_+_15973122 | 0.28 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr23_+_19590006 | 0.28 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr12_+_15002757 | 0.28 |
ENSDART00000135036
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr23_+_9522781 | 0.26 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr15_+_24737599 | 0.23 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr18_-_2549198 | 0.21 |
ENSDART00000186516
|
CABZ01070631.1
|
|
| chr3_-_15144067 | 0.20 |
ENSDART00000127738
ENSDART00000060426 ENSDART00000180799 |
fam173a
|
family with sequence similarity 173, member A |
| chr17_-_31308658 | 0.16 |
ENSDART00000124505
|
bahd1
|
bromo adjacent homology domain containing 1 |
| chr5_-_69437422 | 0.16 |
ENSDART00000073676
|
isca1
|
iron-sulfur cluster assembly 1 |
| chr6_-_14038804 | 0.15 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr13_+_30035253 | 0.15 |
ENSDART00000181303
ENSDART00000057525 ENSDART00000136622 |
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr22_-_10605045 | 0.14 |
ENSDART00000184812
|
bap1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr15_+_21882419 | 0.11 |
ENSDART00000157216
|
si:dkey-103g5.4
|
si:dkey-103g5.4 |
| chr22_+_10606573 | 0.11 |
ENSDART00000192638
|
rad54l2
|
RAD54 like 2 |
| chr8_+_21225064 | 0.10 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr3_+_24094581 | 0.09 |
ENSDART00000138270
ENSDART00000131509 |
copz2
|
coatomer protein complex, subunit zeta 2 |
| chr25_-_18330503 | 0.09 |
ENSDART00000104496
|
dusp6
|
dual specificity phosphatase 6 |
| chr10_-_20523405 | 0.09 |
ENSDART00000114824
|
ddhd2
|
DDHD domain containing 2 |
| chr25_-_12803723 | 0.07 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr4_+_21867522 | 0.07 |
ENSDART00000140400
|
acss3
|
acyl-CoA synthetase short chain family member 3 |
| chr14_+_8174828 | 0.06 |
ENSDART00000167228
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr20_-_20270191 | 0.05 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr5_-_54395488 | 0.04 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr10_-_25591194 | 0.04 |
ENSDART00000131640
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr8_-_37249813 | 0.03 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr20_-_36617313 | 0.01 |
ENSDART00000172395
ENSDART00000152856 |
enah
|
enabled homolog (Drosophila) |
| chr16_-_54800202 | 0.01 |
ENSDART00000059560
ENSDART00000161833 |
khdc4
|
KH domain containing 4, pre-mRNA splicing factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of tefa+tefb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.3 | 1.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.3 | 1.0 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.3 | 0.8 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.2 | 0.7 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 1.1 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.4 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.1 | 2.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.8 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 1.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.8 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 1.6 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 1.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 1.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.5 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 2.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.0 | 0.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.6 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 2.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 1.6 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 1.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.1 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.0 | 0.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 1.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 1.1 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 2.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.9 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.4 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) positive regulation of protein modification by small protein conjugation or removal(GO:1903322) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0007520 | myoblast fusion(GO:0007520) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.2 | 1.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.3 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 2.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 1.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 2.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.4 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 3.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 2.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 3.4 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 2.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.4 | GO:0030424 | axon(GO:0030424) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 1.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 0.9 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 1.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.1 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.5 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.4 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 2.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.8 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 0.4 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 1.0 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.4 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 0.8 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 2.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 3.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.0 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 4.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 4.5 | GO:0015631 | tubulin binding(GO:0015631) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |