Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
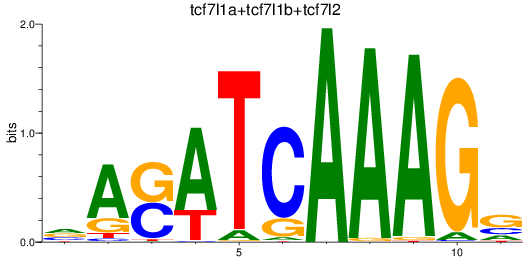
Results for tcf7l1a+tcf7l1b+tcf7l2
Z-value: 2.16
Transcription factors associated with tcf7l1a+tcf7l1b+tcf7l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf7l2
|
ENSDARG00000004415 | transcription factor 7 like 2 |
|
tcf7l1b
|
ENSDARG00000007369 | transcription factor 7 like 1b |
|
tcf7l1a
|
ENSDARG00000038159 | transcription factor 7 like 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf7l2 | dr11_v1_chr12_-_31012741_31012741 | 0.92 | 2.8e-02 | Click! |
| tcf7l1b | dr11_v1_chr8_-_52229462_52229462 | 0.66 | 2.2e-01 | Click! |
| tcf7l1a | dr11_v1_chr10_-_42237304_42237304 | -0.30 | 6.2e-01 | Click! |
Activity profile of tcf7l1a+tcf7l1b+tcf7l2 motif
Sorted Z-values of tcf7l1a+tcf7l1b+tcf7l2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_39011514 | 3.31 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr25_+_21829777 | 1.94 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr13_+_27073901 | 1.84 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr9_+_7548533 | 1.83 |
ENSDART00000081543
|
ptprna
|
protein tyrosine phosphatase, receptor type, Na |
| chr24_+_11105786 | 1.79 |
ENSDART00000175182
|
prlh2
|
prolactin releasing hormone 2 |
| chr4_+_69619 | 1.73 |
ENSDART00000164425
|
mansc1
|
MANSC domain containing 1 |
| chr22_-_21150845 | 1.66 |
ENSDART00000027345
|
tmem59l
|
transmembrane protein 59-like |
| chr2_-_34483597 | 1.64 |
ENSDART00000133224
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr14_+_22172047 | 1.62 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr19_+_31771270 | 1.61 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr15_-_12545683 | 1.61 |
ENSDART00000162807
|
scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr12_+_40905427 | 1.61 |
ENSDART00000170526
ENSDART00000185771 ENSDART00000193945 |
CDH18
|
cadherin 18 |
| chr20_+_22666548 | 1.56 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr8_+_36803415 | 1.51 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr17_+_27176243 | 1.50 |
ENSDART00000162527
|
si:ch211-160f23.7
|
si:ch211-160f23.7 |
| chr1_-_8428736 | 1.49 |
ENSDART00000138435
ENSDART00000121823 |
syngr3b
|
synaptogyrin 3b |
| chr5_+_44190974 | 1.48 |
ENSDART00000182634
ENSDART00000190626 |
TMEM8B
|
si:dkey-84j12.1 |
| chr24_+_32411753 | 1.48 |
ENSDART00000058530
|
neurod6a
|
neuronal differentiation 6a |
| chr16_+_32559821 | 1.46 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr4_+_10366532 | 1.46 |
ENSDART00000189901
|
kcnd2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr1_-_42779075 | 1.44 |
ENSDART00000133917
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr20_+_41549200 | 1.42 |
ENSDART00000135715
|
fam184a
|
family with sequence similarity 184, member A |
| chr7_+_27977065 | 1.42 |
ENSDART00000089574
|
tub
|
tubby bipartite transcription factor |
| chr3_-_19091024 | 1.41 |
ENSDART00000188485
ENSDART00000110554 |
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr1_-_42778510 | 1.39 |
ENSDART00000190172
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr11_+_19068442 | 1.36 |
ENSDART00000171766
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr21_-_25522906 | 1.34 |
ENSDART00000110923
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr13_-_31441042 | 1.34 |
ENSDART00000076571
|
rtn1a
|
reticulon 1a |
| chr25_-_19395476 | 1.32 |
ENSDART00000182622
|
map1ab
|
microtubule-associated protein 1Ab |
| chr23_+_16620801 | 1.30 |
ENSDART00000189859
ENSDART00000184578 |
snphb
|
syntaphilin b |
| chr25_+_7229046 | 1.29 |
ENSDART00000149965
ENSDART00000041820 |
lingo1a
|
leucine rich repeat and Ig domain containing 1a |
| chr8_+_14778292 | 1.29 |
ENSDART00000089971
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr16_+_39159752 | 1.28 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr22_+_3045495 | 1.25 |
ENSDART00000164061
|
LO017843.1
|
|
| chr1_+_32528097 | 1.23 |
ENSDART00000128317
|
nlgn4a
|
neuroligin 4a |
| chr18_-_34143189 | 1.22 |
ENSDART00000079341
|
plch1
|
phospholipase C, eta 1 |
| chr18_+_16330720 | 1.19 |
ENSDART00000080638
|
nts
|
neurotensin |
| chr3_-_30861177 | 1.16 |
ENSDART00000154811
|
shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr16_+_39146696 | 1.15 |
ENSDART00000121756
ENSDART00000084381 |
sybu
|
syntabulin (syntaxin-interacting) |
| chr23_-_26077038 | 1.15 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr18_+_45796096 | 1.13 |
ENSDART00000087070
|
abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr20_+_30445971 | 1.13 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr17_-_20897250 | 1.13 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr15_+_40188076 | 1.12 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr9_-_35155089 | 1.12 |
ENSDART00000077901
|
appb
|
amyloid beta (A4) precursor protein b |
| chr3_-_55650771 | 1.12 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr2_-_30784198 | 1.12 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr1_+_49814461 | 1.10 |
ENSDART00000132405
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr15_-_10341048 | 1.09 |
ENSDART00000171013
|
tenm4
|
teneurin transmembrane protein 4 |
| chr12_+_35203091 | 1.09 |
ENSDART00000153022
|
ndst2b
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2b |
| chr1_-_29045426 | 1.05 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr20_-_40451115 | 1.04 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr5_-_18474486 | 1.03 |
ENSDART00000090580
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr14_+_22591624 | 1.03 |
ENSDART00000108987
|
gfra4b
|
GDNF family receptor alpha 4b |
| chr21_-_37889727 | 1.03 |
ENSDART00000163612
ENSDART00000180958 |
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr25_+_26921480 | 1.03 |
ENSDART00000155949
|
grm8b
|
glutamate receptor, metabotropic 8b |
| chr2_-_22230326 | 1.02 |
ENSDART00000127810
|
fam110b
|
family with sequence similarity 110, member B |
| chr21_-_17482465 | 1.02 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr24_-_24848612 | 1.01 |
ENSDART00000190941
|
crhb
|
corticotropin releasing hormone b |
| chr9_-_37613792 | 0.98 |
ENSDART00000138345
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr24_-_32408404 | 0.98 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr17_-_8638713 | 0.98 |
ENSDART00000148971
|
ctbp2a
|
C-terminal binding protein 2a |
| chr5_-_22082918 | 0.98 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr8_-_50346246 | 0.97 |
ENSDART00000025008
|
stc1
|
stanniocalcin 1 |
| chr4_-_77432218 | 0.97 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr6_-_53143667 | 0.96 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr9_-_12034444 | 0.95 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr12_-_31103187 | 0.94 |
ENSDART00000005562
ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 ENSDART00000122972 ENSDART00000153068 |
tcf7l2
|
transcription factor 7 like 2 |
| chr21_-_25522510 | 0.93 |
ENSDART00000162711
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr6_-_26559921 | 0.93 |
ENSDART00000104532
|
sox14
|
SRY (sex determining region Y)-box 14 |
| chr3_-_55650417 | 0.92 |
ENSDART00000171441
|
axin2
|
axin 2 (conductin, axil) |
| chr17_-_4245311 | 0.92 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr11_+_34522554 | 0.91 |
ENSDART00000109833
|
zmat3
|
zinc finger, matrin-type 3 |
| chr8_-_34051548 | 0.91 |
ENSDART00000105204
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr6_-_15604157 | 0.90 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr2_-_30784502 | 0.90 |
ENSDART00000056735
|
rgs20
|
regulator of G protein signaling 20 |
| chr23_-_20764227 | 0.90 |
ENSDART00000089750
|
znf362b
|
zinc finger protein 362b |
| chr9_-_3671911 | 0.89 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr9_+_32431666 | 0.88 |
ENSDART00000187614
|
plcl1
|
phospholipase C like 1 |
| chr16_-_18702249 | 0.87 |
ENSDART00000191595
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr6_-_29105727 | 0.86 |
ENSDART00000184355
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr3_-_2589704 | 0.84 |
ENSDART00000187239
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr14_-_47963115 | 0.84 |
ENSDART00000003826
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr24_+_15897717 | 0.81 |
ENSDART00000105956
|
neto1l
|
neuropilin (NRP) and tolloid (TLL)-like 1, like |
| chr15_+_22722684 | 0.80 |
ENSDART00000156760
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr18_+_36806545 | 0.80 |
ENSDART00000098958
|
ttc9b
|
tetratricopeptide repeat domain 9B |
| chr20_+_36730049 | 0.79 |
ENSDART00000045948
|
ncoa1
|
nuclear receptor coactivator 1 |
| chr16_+_34493987 | 0.79 |
ENSDART00000138374
|
si:ch211-255i3.4
|
si:ch211-255i3.4 |
| chr10_-_31562695 | 0.79 |
ENSDART00000186456
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr3_-_33573252 | 0.79 |
ENSDART00000191339
ENSDART00000184612 ENSDART00000187232 ENSDART00000190671 |
CR847537.1
|
|
| chr20_-_9436521 | 0.79 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr22_-_16042243 | 0.78 |
ENSDART00000062633
|
s1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr12_+_10115964 | 0.77 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr2_+_9822319 | 0.76 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr11_+_34523132 | 0.76 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr2_-_43083749 | 0.72 |
ENSDART00000084303
ENSDART00000145494 |
kcnq3
|
potassium voltage-gated channel, KQT-like subfamily, member 3 |
| chr20_+_36234335 | 0.72 |
ENSDART00000193484
ENSDART00000181664 |
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr8_-_22739757 | 0.72 |
ENSDART00000182167
ENSDART00000171891 |
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr3_-_32299892 | 0.72 |
ENSDART00000181472
|
si:dkey-16p21.7
|
si:dkey-16p21.7 |
| chr18_+_10907999 | 0.71 |
ENSDART00000133555
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr16_+_17389116 | 0.71 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr15_-_2188332 | 0.71 |
ENSDART00000138941
ENSDART00000009564 |
shox2
|
short stature homeobox 2 |
| chr17_+_26611929 | 0.70 |
ENSDART00000166450
ENSDART00000087023 |
ttc7b
|
tetratricopeptide repeat domain 7B |
| chr15_+_24691088 | 0.70 |
ENSDART00000110618
|
LRRC75A
|
si:dkey-151p21.7 |
| chr21_-_31241254 | 0.69 |
ENSDART00000065365
|
tpst1l
|
tyrosylprotein sulfotransferase 1, like |
| chr15_-_163586 | 0.69 |
ENSDART00000163597
|
SEPT4
|
septin-4 |
| chr13_+_4505232 | 0.68 |
ENSDART00000007500
ENSDART00000161684 |
pde10a
|
phosphodiesterase 10A |
| chr19_+_29798064 | 0.68 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr8_-_34052019 | 0.68 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr1_-_36151377 | 0.67 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr6_-_57938043 | 0.67 |
ENSDART00000171073
|
tox2
|
TOX high mobility group box family member 2 |
| chr1_+_2190714 | 0.66 |
ENSDART00000132126
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr7_-_42206720 | 0.66 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr1_-_39976492 | 0.66 |
ENSDART00000181680
|
stox2a
|
storkhead box 2a |
| chr17_-_4245902 | 0.65 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr7_-_37622153 | 0.64 |
ENSDART00000052379
|
nkd1
|
naked cuticle homolog 1 (Drosophila) |
| chr7_-_37622370 | 0.64 |
ENSDART00000173523
|
nkd1
|
naked cuticle homolog 1 (Drosophila) |
| chr21_-_25722834 | 0.64 |
ENSDART00000101208
|
abhd11
|
abhydrolase domain containing 11 |
| chr13_+_15656042 | 0.64 |
ENSDART00000134240
|
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr24_-_6546479 | 0.63 |
ENSDART00000160538
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr1_+_49814942 | 0.63 |
ENSDART00000164936
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr6_-_7208119 | 0.62 |
ENSDART00000105148
ENSDART00000185846 |
hs6st3a
|
heparan sulfate 6-O-sulfotransferase 3a |
| chr17_-_24714837 | 0.61 |
ENSDART00000154871
|
si:ch211-15d5.11
|
si:ch211-15d5.11 |
| chr6_+_15762647 | 0.60 |
ENSDART00000127133
ENSDART00000128939 |
iqca1
|
IQ motif containing with AAA domain 1 |
| chr5_+_15992655 | 0.60 |
ENSDART00000182148
|
znrf3
|
zinc and ring finger 3 |
| chr12_+_48340133 | 0.60 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr8_+_17009199 | 0.60 |
ENSDART00000139452
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr9_+_28693592 | 0.60 |
ENSDART00000110198
|
zgc:162780
|
zgc:162780 |
| chr19_-_8880688 | 0.59 |
ENSDART00000039629
|
celf3a
|
cugbp, Elav-like family member 3a |
| chr6_-_53144336 | 0.59 |
ENSDART00000154429
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr7_-_31940590 | 0.58 |
ENSDART00000131009
|
bdnf
|
brain-derived neurotrophic factor |
| chr25_-_19395156 | 0.57 |
ENSDART00000155335
|
map1ab
|
microtubule-associated protein 1Ab |
| chr19_-_8732037 | 0.57 |
ENSDART00000138971
|
si:ch211-39a7.1
|
si:ch211-39a7.1 |
| chr3_+_15809098 | 0.56 |
ENSDART00000183023
|
phospho1
|
phosphatase, orphan 1 |
| chr3_+_15773991 | 0.55 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr15_-_31588162 | 0.55 |
ENSDART00000153598
|
hsph1
|
heat shock 105/110 protein 1 |
| chr14_+_34495216 | 0.54 |
ENSDART00000147756
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr2_+_33368414 | 0.54 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr17_-_50010121 | 0.53 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr13_-_24906307 | 0.53 |
ENSDART00000148191
ENSDART00000189810 |
kat6b
|
K(lysine) acetyltransferase 6B |
| chr4_-_16658514 | 0.52 |
ENSDART00000133837
|
dennd5b
|
DENN/MADD domain containing 5B |
| chr21_-_40782393 | 0.52 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr12_-_31995840 | 0.52 |
ENSDART00000112881
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr22_+_35930526 | 0.52 |
ENSDART00000169242
|
LO016987.1
|
|
| chr1_+_26356360 | 0.51 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr15_+_28106498 | 0.51 |
ENSDART00000041707
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr1_+_26356205 | 0.51 |
ENSDART00000190064
ENSDART00000176380 |
tet2
|
tet methylcytosine dioxygenase 2 |
| chr16_+_16933002 | 0.50 |
ENSDART00000173163
|
myh14
|
myosin, heavy chain 14, non-muscle |
| chr9_+_10014817 | 0.50 |
ENSDART00000132065
|
nxph2a
|
neurexophilin 2a |
| chr2_-_42396592 | 0.50 |
ENSDART00000127136
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr6_-_21616659 | 0.49 |
ENSDART00000074256
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr19_-_44801918 | 0.49 |
ENSDART00000108507
|
CSMD3 (1 of many)
|
si:ch211-233f16.1 |
| chr21_+_25068215 | 0.49 |
ENSDART00000167523
ENSDART00000189259 |
dixdc1b
|
DIX domain containing 1b |
| chr23_+_25201077 | 0.49 |
ENSDART00000136675
|
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr6_-_15603675 | 0.48 |
ENSDART00000143502
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr13_+_4505079 | 0.48 |
ENSDART00000144312
|
pde10a
|
phosphodiesterase 10A |
| chr19_-_23895839 | 0.47 |
ENSDART00000174834
|
si:dkey-222b8.4
|
si:dkey-222b8.4 |
| chr17_-_26507289 | 0.46 |
ENSDART00000155616
|
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr17_+_3379673 | 0.46 |
ENSDART00000176354
|
sntg2
|
syntrophin, gamma 2 |
| chr1_-_19233890 | 0.46 |
ENSDART00000127145
|
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr19_-_32940040 | 0.46 |
ENSDART00000179947
|
azin1b
|
antizyme inhibitor 1b |
| chr7_+_27976448 | 0.46 |
ENSDART00000181026
|
tub
|
tubby bipartite transcription factor |
| chr5_+_34407763 | 0.46 |
ENSDART00000188849
ENSDART00000145127 |
lamc3
|
laminin, gamma 3 |
| chr20_+_43942278 | 0.46 |
ENSDART00000100571
|
clic5b
|
chloride intracellular channel 5b |
| chr7_+_57836841 | 0.45 |
ENSDART00000136175
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr11_+_14622379 | 0.45 |
ENSDART00000112589
|
efna2b
|
ephrin-A2b |
| chr17_+_26352372 | 0.45 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr13_-_18835254 | 0.43 |
ENSDART00000147579
ENSDART00000146795 |
lzts2a
|
leucine zipper, putative tumor suppressor 2a |
| chr7_-_34062301 | 0.43 |
ENSDART00000052404
|
map2k5
|
mitogen-activated protein kinase kinase 5 |
| chr15_-_43768776 | 0.43 |
ENSDART00000170398
|
grm5b
|
glutamate receptor, metabotropic 5b |
| chr20_-_9194257 | 0.43 |
ENSDART00000133012
|
ylpm1
|
YLP motif containing 1 |
| chr7_-_72423666 | 0.41 |
ENSDART00000191214
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr14_+_24277556 | 0.41 |
ENSDART00000122660
|
hnrnpa0a
|
heterogeneous nuclear ribonucleoprotein A0a |
| chr9_+_10014514 | 0.41 |
ENSDART00000185590
|
nxph2a
|
neurexophilin 2a |
| chr15_+_1705167 | 0.40 |
ENSDART00000081940
|
otol1b
|
otolin 1b |
| chr7_-_32895668 | 0.40 |
ENSDART00000141828
|
ano5b
|
anoctamin 5b |
| chr5_+_26156079 | 0.40 |
ENSDART00000088141
|
ankrd34bb
|
ankyrin repeat domain 34Bb |
| chr7_-_31941330 | 0.40 |
ENSDART00000144682
|
bdnf
|
brain-derived neurotrophic factor |
| chr17_-_20897407 | 0.39 |
ENSDART00000149481
|
ank3b
|
ankyrin 3b |
| chr11_-_18604317 | 0.39 |
ENSDART00000182081
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr18_-_44888375 | 0.39 |
ENSDART00000160506
|
si:ch211-71n6.4
|
si:ch211-71n6.4 |
| chr15_+_6109861 | 0.38 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr11_-_18323059 | 0.37 |
ENSDART00000182590
|
SFMBT1
|
Scm like with four mbt domains 1 |
| chr24_-_35269991 | 0.36 |
ENSDART00000185424
|
sntg1
|
syntrophin, gamma 1 |
| chr4_-_2036620 | 0.35 |
ENSDART00000150490
|
si:dkey-97m3.1
|
si:dkey-97m3.1 |
| chr6_-_14038804 | 0.35 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr11_+_28107632 | 0.33 |
ENSDART00000177721
|
nmur3
|
neuromedin U receptor 3 |
| chr15_-_16384184 | 0.33 |
ENSDART00000154504
|
fam222bb
|
family with sequence similarity 222, member Bb |
| chr2_+_2503396 | 0.33 |
ENSDART00000168418
|
crhr2
|
corticotropin releasing hormone receptor 2 |
| chr2_+_22042745 | 0.33 |
ENSDART00000132039
|
tox
|
thymocyte selection-associated high mobility group box |
| chr11_+_41135055 | 0.32 |
ENSDART00000173252
|
camta1
|
calmodulin binding transcription activator 1 |
| chr3_+_28831622 | 0.32 |
ENSDART00000184130
ENSDART00000191294 |
flr
|
fleer |
| chr23_-_24226533 | 0.32 |
ENSDART00000109134
|
plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr13_+_28702104 | 0.32 |
ENSDART00000135481
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr6_-_54111928 | 0.31 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr12_+_32323098 | 0.31 |
ENSDART00000188722
|
ANKFN1
|
si:ch211-277e21.2 |
| chr4_-_20511595 | 0.31 |
ENSDART00000185806
|
rassf8b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8b |
| chr13_-_23264724 | 0.31 |
ENSDART00000051886
|
si:dkey-103j14.5
|
si:dkey-103j14.5 |
| chr7_+_42206543 | 0.30 |
ENSDART00000112543
|
phkb
|
phosphorylase kinase, beta |
| chr9_-_27410597 | 0.30 |
ENSDART00000135652
ENSDART00000042297 |
kdelc1
|
KDEL (Lys-Asp-Glu-Leu) containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf7l1a+tcf7l1b+tcf7l2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.4 | 1.1 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.3 | 1.7 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.3 | 1.0 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.3 | 1.3 | GO:0042306 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.3 | 1.6 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.3 | 0.8 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.3 | 1.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.2 | 0.9 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.2 | 1.2 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.2 | 1.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.2 | 1.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 0.5 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.2 | 1.5 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.2 | 1.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 2.0 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.2 | 0.6 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 1.9 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 1.0 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.1 | 0.4 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 1.0 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.5 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 1.1 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 2.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.1 | 0.9 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 1.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 1.0 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 1.0 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.7 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 1.0 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.3 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 0.5 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 1.9 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.4 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.5 | GO:1900186 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.2 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 2.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.3 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.2 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.0 | 1.9 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 1.0 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.8 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.5 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 1.3 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.3 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 1.0 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.3 | GO:1903729 | regulation of plasma membrane organization(GO:1903729) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:1903318 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 0.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 1.0 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.8 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.2 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.1 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.4 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.6 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.4 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 1.3 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 2.8 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 1.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 1.5 | GO:0050769 | positive regulation of neurogenesis(GO:0050769) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.2 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.3 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.2 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.0 | 0.7 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 3.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.1 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.0 | 0.2 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 1.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.8 | GO:0030042 | actin filament depolymerization(GO:0030042) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 0.8 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 0.5 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 2.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 1.6 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.4 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 2.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 1.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 5.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 1.0 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.9 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 1.3 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.2 | GO:0030133 | transport vesicle(GO:0030133) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.2 | 1.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 1.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 1.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 1.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 0.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.9 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 1.0 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 1.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 1.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 2.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.3 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.1 | 1.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 2.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.3 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 0.6 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.8 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 1.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.7 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.3 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.5 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 1.0 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 0.2 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.3 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.4 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 2.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.2 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.1 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 1.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 5.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.4 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 3.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 3.8 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.0 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 2.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 1.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 0.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 2.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 2.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |