Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
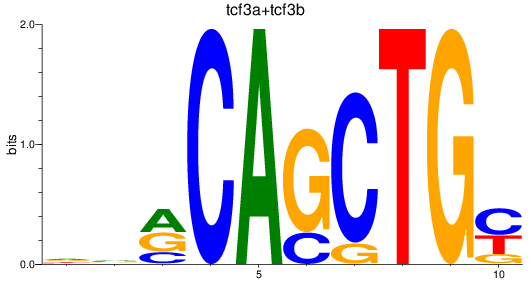
Results for tcf3a+tcf3b
Z-value: 1.62
Transcription factors associated with tcf3a+tcf3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf3a
|
ENSDARG00000005915 | transcription factor 3a |
|
tcf3b
|
ENSDARG00000099999 | transcription factor 3b |
|
tcf3b
|
ENSDARG00000112646 | transcription factor 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf3a | dr11_v1_chr2_-_57378748_57378748 | -0.55 | 3.3e-01 | Click! |
| tcf3b | dr11_v1_chr22_-_20342260_20342260 | -0.03 | 9.6e-01 | Click! |
Activity profile of tcf3a+tcf3b motif
Sorted Z-values of tcf3a+tcf3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_11756040 | 1.71 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr22_+_11775269 | 1.01 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr23_-_5683147 | 0.93 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr8_+_1009831 | 0.79 |
ENSDART00000172414
|
fabp1b.2
|
fatty acid binding protein 1b, liver, tandem duplicate 2 |
| chr23_+_25893020 | 0.75 |
ENSDART00000144769
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr13_-_438705 | 0.74 |
ENSDART00000082142
|
CU570800.1
|
|
| chr14_+_31651533 | 0.72 |
ENSDART00000172835
|
fhl1a
|
four and a half LIM domains 1a |
| chr22_-_10541372 | 0.71 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr7_+_31838320 | 0.70 |
ENSDART00000144679
ENSDART00000174217 ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr4_-_4834347 | 0.69 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr2_-_42128714 | 0.69 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr16_+_813780 | 0.68 |
ENSDART00000162474
ENSDART00000161774 |
irx1a
|
iroquois homeobox 1a |
| chr23_-_10177442 | 0.66 |
ENSDART00000144280
ENSDART00000129044 |
krt5
|
keratin 5 |
| chr8_-_1051438 | 0.66 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr22_+_16308806 | 0.65 |
ENSDART00000162685
|
lrrc39
|
leucine rich repeat containing 39 |
| chr3_-_29962345 | 0.63 |
ENSDART00000136819
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr20_-_9436521 | 0.62 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr6_-_35439406 | 0.61 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr22_+_16308450 | 0.59 |
ENSDART00000105678
|
lrrc39
|
leucine rich repeat containing 39 |
| chr6_-_42003780 | 0.58 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr7_+_31891110 | 0.58 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr19_-_11336782 | 0.58 |
ENSDART00000131014
|
sept7a
|
septin 7a |
| chr18_-_14337065 | 0.57 |
ENSDART00000135703
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr1_+_55137943 | 0.55 |
ENSDART00000138070
ENSDART00000150510 ENSDART00000133472 ENSDART00000136378 |
mb
|
myoglobin |
| chr6_-_35401282 | 0.55 |
ENSDART00000127612
|
rgs5a
|
regulator of G protein signaling 5a |
| chr9_-_100579 | 0.55 |
ENSDART00000006099
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr21_+_13205859 | 0.54 |
ENSDART00000102253
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr14_+_31657412 | 0.54 |
ENSDART00000105767
|
fhl1a
|
four and a half LIM domains 1a |
| chr23_+_25879320 | 0.54 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr6_-_15653494 | 0.53 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr2_+_55982940 | 0.52 |
ENSDART00000097753
ENSDART00000097751 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr7_-_35708450 | 0.52 |
ENSDART00000193886
|
irx5a
|
iroquois homeobox 5a |
| chr23_+_19790962 | 0.51 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr18_-_6634424 | 0.51 |
ENSDART00000062423
ENSDART00000179955 |
tnni1c
|
troponin I, skeletal, slow c |
| chr20_+_46040666 | 0.50 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr18_+_5454341 | 0.50 |
ENSDART00000192649
|
dtwd1
|
DTW domain containing 1 |
| chr5_+_1911814 | 0.49 |
ENSDART00000172233
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr3_-_34095221 | 0.49 |
ENSDART00000164235
ENSDART00000151377 |
ighv1-4
ighv5-4
|
immunoglobulin heavy variable 1-4 immunoglobulin heavy variable 5-4 |
| chr18_+_2228737 | 0.49 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr22_-_10541712 | 0.49 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr5_-_26118855 | 0.48 |
ENSDART00000009028
|
ela3l
|
elastase 3 like |
| chr22_-_651719 | 0.48 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr22_-_57177 | 0.48 |
ENSDART00000163959
|
CABZ01085139.1
|
|
| chr11_-_18705303 | 0.47 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr12_-_26064105 | 0.45 |
ENSDART00000168825
|
ldb3b
|
LIM domain binding 3b |
| chr5_-_36837846 | 0.45 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr18_-_49020066 | 0.45 |
ENSDART00000174394
|
BX663503.3
|
|
| chr17_+_1323699 | 0.45 |
ENSDART00000172540
|
adssl1
|
adenylosuccinate synthase like 1 |
| chr3_-_50865079 | 0.44 |
ENSDART00000164295
|
pmp22a
|
peripheral myelin protein 22a |
| chr7_+_65586624 | 0.44 |
ENSDART00000184344
|
mical2a
|
microtubule associated monooxygenase, calponin and LIM domain containing 2a |
| chr8_+_21376290 | 0.44 |
ENSDART00000136765
|
ela2
|
elastase 2 |
| chr17_-_681142 | 0.44 |
ENSDART00000165583
|
soul3
|
heme-binding protein soul3 |
| chr3_-_30685401 | 0.44 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr7_-_44963154 | 0.43 |
ENSDART00000073735
|
rrad
|
Ras-related associated with diabetes |
| chr24_-_9300160 | 0.43 |
ENSDART00000152378
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr17_+_26657896 | 0.43 |
ENSDART00000152037
|
ttc7b
|
tetratricopeptide repeat domain 7B |
| chr19_-_5372572 | 0.43 |
ENSDART00000151326
|
krt17
|
keratin 17 |
| chr7_-_48263516 | 0.42 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr16_+_26012569 | 0.42 |
ENSDART00000148846
|
prss59.1
|
protease, serine, 59, tandem duplicate 1 |
| chr13_+_23157053 | 0.42 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr8_-_4586696 | 0.42 |
ENSDART00000143511
ENSDART00000134324 |
gp1bb
|
glycoprotein Ib (platelet), beta polypeptide |
| chr4_-_14915268 | 0.42 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr6_-_21189295 | 0.42 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr9_-_98982 | 0.42 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr4_-_20181964 | 0.41 |
ENSDART00000022539
|
fgl2a
|
fibrinogen-like 2a |
| chr1_-_47431453 | 0.41 |
ENSDART00000101104
|
gja5b
|
gap junction protein, alpha 5b |
| chr1_-_156375 | 0.41 |
ENSDART00000160221
|
pcid2
|
PCI domain containing 2 |
| chr20_+_23501535 | 0.40 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr3_+_59935606 | 0.40 |
ENSDART00000154157
|
arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr7_+_35229805 | 0.40 |
ENSDART00000173911
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr22_-_27115241 | 0.40 |
ENSDART00000019442
|
xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr1_+_38142354 | 0.40 |
ENSDART00000179352
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr11_+_24313931 | 0.40 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr11_+_37096520 | 0.40 |
ENSDART00000181503
|
card19
|
caspase recruitment domain family, member 19 |
| chr17_+_38573471 | 0.39 |
ENSDART00000040627
|
sptb
|
spectrin, beta, erythrocytic |
| chr2_-_21352101 | 0.39 |
ENSDART00000057021
|
hhatla
|
hedgehog acyltransferase like, a |
| chr11_-_28050559 | 0.39 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr2_+_55982300 | 0.38 |
ENSDART00000183903
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr25_-_3867990 | 0.38 |
ENSDART00000075663
|
cracr2b
|
calcium release activated channel regulator 2B |
| chr14_-_25956804 | 0.38 |
ENSDART00000135627
ENSDART00000146022 ENSDART00000039660 |
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr18_+_29145681 | 0.38 |
ENSDART00000089031
ENSDART00000193336 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr24_+_39108243 | 0.37 |
ENSDART00000156353
|
mss51
|
MSS51 mitochondrial translational activator |
| chr23_+_25708787 | 0.37 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr1_-_38816685 | 0.37 |
ENSDART00000075230
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr12_-_28570989 | 0.37 |
ENSDART00000008010
|
pdk2a
|
pyruvate dehydrogenase kinase, isozyme 2a |
| chr14_+_146857 | 0.37 |
ENSDART00000122521
|
CABZ01088229.1
|
|
| chr17_-_25382367 | 0.37 |
ENSDART00000162306
ENSDART00000165282 |
lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr13_+_52061034 | 0.37 |
ENSDART00000170383
|
CABZ01089777.1
|
|
| chr1_+_1915967 | 0.37 |
ENSDART00000131463
|
si:ch211-132g1.1
|
si:ch211-132g1.1 |
| chr11_+_24314148 | 0.36 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr13_-_349952 | 0.36 |
ENSDART00000133731
ENSDART00000140326 ENSDART00000189389 ENSDART00000185865 ENSDART00000109634 ENSDART00000147058 ENSDART00000142695 |
si:ch1073-291c23.2
|
si:ch1073-291c23.2 |
| chr18_+_26899316 | 0.36 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr6_-_39051319 | 0.36 |
ENSDART00000155093
|
tns2b
|
tensin 2b |
| chr5_-_26566435 | 0.35 |
ENSDART00000146070
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr7_-_41858513 | 0.35 |
ENSDART00000109918
|
mylk3
|
myosin light chain kinase 3 |
| chr14_-_17563773 | 0.35 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor like 1a |
| chr5_-_22052852 | 0.35 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr16_-_31469065 | 0.35 |
ENSDART00000182397
|
si:ch211-251p5.5
|
si:ch211-251p5.5 |
| chr7_+_22767678 | 0.35 |
ENSDART00000137203
|
ponzr6
|
plac8 onzin related protein 6 |
| chr8_-_18582922 | 0.35 |
ENSDART00000123917
|
tmem47
|
transmembrane protein 47 |
| chr21_+_28747069 | 0.35 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr21_+_28747236 | 0.34 |
ENSDART00000137874
|
zgc:100829
|
zgc:100829 |
| chr23_+_18722715 | 0.34 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr19_+_46113828 | 0.34 |
ENSDART00000159331
ENSDART00000161826 |
rbm24a
|
RNA binding motif protein 24a |
| chr23_+_18722915 | 0.33 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr24_+_8736497 | 0.33 |
ENSDART00000181904
|
tmem14ca
|
transmembrane protein 14Ca |
| chr17_+_32360673 | 0.33 |
ENSDART00000155519
|
si:ch211-139d20.3
|
si:ch211-139d20.3 |
| chr10_-_35410518 | 0.32 |
ENSDART00000048430
|
gabrr3a
|
gamma-aminobutyric acid (GABA) A receptor, rho 3a |
| chr23_+_19213472 | 0.32 |
ENSDART00000185985
|
apobec2b
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2b |
| chr18_-_14337450 | 0.32 |
ENSDART00000061435
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr8_-_7093507 | 0.32 |
ENSDART00000045669
|
si:dkey-222n6.2
|
si:dkey-222n6.2 |
| chr2_+_45643122 | 0.32 |
ENSDART00000138179
|
fndc7rs4
|
fibronectin type III domain containing 7, related sequence 4 |
| chr20_-_26551210 | 0.32 |
ENSDART00000077715
|
si:dkey-25e12.3
|
si:dkey-25e12.3 |
| chr21_+_28749720 | 0.31 |
ENSDART00000145178
|
zgc:100829
|
zgc:100829 |
| chr1_-_59567685 | 0.31 |
ENSDART00000159144
ENSDART00000182913 |
zmp:0000001082
|
zmp:0000001082 |
| chr19_+_31904836 | 0.31 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr3_+_13559199 | 0.31 |
ENSDART00000166547
|
si:ch73-106n3.1
|
si:ch73-106n3.1 |
| chr18_-_26101800 | 0.31 |
ENSDART00000004692
|
idh2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr7_-_73843720 | 0.31 |
ENSDART00000111622
|
caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr22_+_7439186 | 0.31 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr13_-_37615029 | 0.31 |
ENSDART00000111199
|
si:dkey-188i13.6
|
si:dkey-188i13.6 |
| chr10_-_35220285 | 0.30 |
ENSDART00000180439
|
ypel2a
|
yippee-like 2a |
| chr5_+_64900223 | 0.30 |
ENSDART00000191677
|
ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr16_+_44768361 | 0.30 |
ENSDART00000036302
|
upk1a
|
uroplakin 1a |
| chr21_-_20840714 | 0.30 |
ENSDART00000144861
ENSDART00000139430 |
c6
|
complement component 6 |
| chr12_+_5708400 | 0.30 |
ENSDART00000017191
|
dlx3b
|
distal-less homeobox 3b |
| chr13_+_25200105 | 0.30 |
ENSDART00000039640
|
ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr12_-_25380028 | 0.30 |
ENSDART00000142674
|
zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr11_-_544187 | 0.30 |
ENSDART00000173161
|
CU695207.1
|
|
| chr13_+_32148338 | 0.29 |
ENSDART00000188591
|
osr1
|
odd-skipped related transciption factor 1 |
| chr25_-_20258508 | 0.29 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr13_+_228045 | 0.29 |
ENSDART00000161091
|
zgc:64201
|
zgc:64201 |
| chr14_-_15699528 | 0.29 |
ENSDART00000161123
|
neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr19_+_24488403 | 0.29 |
ENSDART00000052421
|
txnipa
|
thioredoxin interacting protein a |
| chr3_+_27713610 | 0.29 |
ENSDART00000019004
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr6_+_2097690 | 0.29 |
ENSDART00000193770
|
tgm2b
|
transglutaminase 2b |
| chr3_-_34060153 | 0.29 |
ENSDART00000151610
|
ighv1-1
|
immunoglobulin heavy variable 1-1 |
| chr21_+_30950097 | 0.29 |
ENSDART00000187572
|
rhogb
|
ras homolog family member Gb |
| chr21_-_4250682 | 0.29 |
ENSDART00000099389
|
dnlz
|
DNL-type zinc finger |
| chr9_-_105135 | 0.28 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr3_-_34069637 | 0.28 |
ENSDART00000151588
|
ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr21_-_45882643 | 0.28 |
ENSDART00000168703
|
galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr13_+_32144370 | 0.28 |
ENSDART00000020270
|
osr1
|
odd-skipped related transciption factor 1 |
| chr11_+_13166750 | 0.28 |
ENSDART00000169961
|
mob3c
|
MOB kinase activator 3C |
| chr23_-_30041065 | 0.28 |
ENSDART00000131209
ENSDART00000127192 |
ccdc187
|
coiled-coil domain containing 187 |
| chr11_-_30636163 | 0.28 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr23_+_19814371 | 0.28 |
ENSDART00000182897
|
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr6_-_10912424 | 0.28 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr3_-_180860 | 0.28 |
ENSDART00000059956
ENSDART00000192506 |
kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr13_+_42124566 | 0.28 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr19_-_43639331 | 0.28 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr21_+_11776328 | 0.28 |
ENSDART00000134469
ENSDART00000081646 |
glrx
|
glutaredoxin (thioltransferase) |
| chr8_-_44298964 | 0.27 |
ENSDART00000098520
|
fzd10
|
frizzled class receptor 10 |
| chr12_-_48168135 | 0.27 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr15_-_29387446 | 0.27 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr10_-_342564 | 0.27 |
ENSDART00000157633
|
vaspa
|
vasodilator stimulated phosphoprotein a |
| chr23_-_31512496 | 0.27 |
ENSDART00000158755
ENSDART00000143425 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr14_+_38786298 | 0.27 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr11_-_43104475 | 0.27 |
ENSDART00000125368
|
acyp2
|
acylphosphatase 2, muscle type |
| chr25_+_16601839 | 0.27 |
ENSDART00000008986
|
atp6v1e1a
|
ATPase H+ transporting V1 subunit E1a |
| chr5_-_26466169 | 0.27 |
ENSDART00000144035
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr3_-_32590164 | 0.27 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr2_-_44255537 | 0.27 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr3_-_4574372 | 0.27 |
ENSDART00000172023
ENSDART00000168724 |
ftr50
|
finTRIM family, member 50 |
| chr15_-_4528326 | 0.27 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr14_+_30340251 | 0.27 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr2_+_22851832 | 0.27 |
ENSDART00000145944
|
amotl2b
|
angiomotin like 2b |
| chr8_-_30242706 | 0.26 |
ENSDART00000139864
ENSDART00000143809 |
zgc:162939
|
zgc:162939 |
| chr5_+_37854685 | 0.26 |
ENSDART00000051222
ENSDART00000185283 |
ins
|
preproinsulin |
| chr7_+_35229645 | 0.26 |
ENSDART00000144327
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr9_-_48281941 | 0.26 |
ENSDART00000099787
|
klhl41a
|
kelch-like family member 41a |
| chr20_+_46385907 | 0.26 |
ENSDART00000060710
|
adgrg11
|
adhesion G protein-coupled receptor G11 |
| chr6_-_48082525 | 0.26 |
ENSDART00000192049
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr20_+_2039518 | 0.26 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr5_+_24287927 | 0.26 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr9_+_30585552 | 0.26 |
ENSDART00000160590
ENSDART00000189815 ENSDART00000143766 |
tbc1d4
|
TBC1 domain family, member 4 |
| chr14_-_36412473 | 0.26 |
ENSDART00000128244
ENSDART00000138376 |
asb5a
|
ankyrin repeat and SOCS box containing 5a |
| chr21_+_5974590 | 0.26 |
ENSDART00000098499
|
dck
|
deoxycytidine kinase |
| chr3_-_34089310 | 0.26 |
ENSDART00000151774
|
ighv5-5
|
immunoglobulin heavy variable 5-5 |
| chr18_+_35842933 | 0.25 |
ENSDART00000151587
ENSDART00000131121 |
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr4_-_16412084 | 0.25 |
ENSDART00000188460
|
dcn
|
decorin |
| chr17_+_132555 | 0.25 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr6_-_10788065 | 0.25 |
ENSDART00000190968
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr17_+_16090436 | 0.25 |
ENSDART00000136059
ENSDART00000138734 |
znf395a
|
zinc finger protein 395a |
| chr24_-_14711597 | 0.25 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr9_-_23990416 | 0.25 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr16_+_3004422 | 0.25 |
ENSDART00000189969
|
CABZ01043952.1
|
|
| chr16_-_21810668 | 0.25 |
ENSDART00000156575
|
dicp3.3
|
diverse immunoglobulin domain-containing protein 3.3 |
| chr9_+_40874194 | 0.25 |
ENSDART00000141548
|
hibch
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr16_-_7239457 | 0.24 |
ENSDART00000148992
ENSDART00000149260 |
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr1_-_17587552 | 0.24 |
ENSDART00000039917
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr20_-_37831849 | 0.24 |
ENSDART00000188483
ENSDART00000153005 ENSDART00000142364 |
si:ch211-147d7.5
|
si:ch211-147d7.5 |
| chr13_-_15994419 | 0.24 |
ENSDART00000079724
ENSDART00000042377 ENSDART00000046079 ENSDART00000050481 ENSDART00000016430 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr15_-_23814330 | 0.24 |
ENSDART00000153843
|
si:ch211-167j9.5
|
si:ch211-167j9.5 |
| chr18_+_25752592 | 0.24 |
ENSDART00000111767
|
si:ch211-39k3.2
|
si:ch211-39k3.2 |
| chr5_-_14521500 | 0.24 |
ENSDART00000176565
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr8_+_50946379 | 0.24 |
ENSDART00000139649
|
b2ml
|
beta-2-microglobulin, like |
| chr2_+_16191852 | 0.24 |
ENSDART00000152046
|
sh3glb1b
|
SH3-domain GRB2-like endophilin B1b |
| chr17_-_45370200 | 0.24 |
ENSDART00000186208
|
znf106a
|
zinc finger protein 106a |
| chr23_-_18130264 | 0.24 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf3a+tcf3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.3 | 0.8 | GO:0043388 | cytidine to uridine editing(GO:0016554) positive regulation of DNA binding(GO:0043388) |
| 0.3 | 1.3 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.2 | 1.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 1.3 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.2 | 0.6 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.2 | 0.9 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.5 | GO:0060254 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.2 | 0.5 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 0.6 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 1.0 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.3 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.8 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.3 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.4 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.2 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.1 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.3 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.5 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 1.0 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 0.2 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.2 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.1 | 0.2 | GO:0098801 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) regulation of renal system process(GO:0098801) |
| 0.1 | 0.2 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 0.2 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.1 | 1.2 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 0.2 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 0.5 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 0.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.3 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.0 | 0.2 | GO:0060406 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.0 | 0.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.0 | 0.5 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 0.0 | 0.2 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.5 | GO:2000403 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.0 | 2.3 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.1 | GO:1990120 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.2 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.3 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.7 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.3 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.0 | 0.2 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.3 | GO:0035912 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.2 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:1900157 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.0 | 0.1 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.0 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.7 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.0 | 0.1 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.2 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.1 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.0 | 0.3 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.4 | GO:0030534 | adult behavior(GO:0030534) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.0 | 0.2 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.1 | GO:0043300 | mast cell activation involved in immune response(GO:0002279) mast cell mediated immunity(GO:0002448) regulation of mast cell activation(GO:0033003) regulation of mast cell activation involved in immune response(GO:0033006) leukocyte degranulation(GO:0043299) regulation of leukocyte degranulation(GO:0043300) mast cell degranulation(GO:0043303) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.2 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.2 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.3 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0051103 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.0 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.8 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 1.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.2 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.1 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.2 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.2 | GO:0019320 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) hexose catabolic process(GO:0019320) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.2 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.0 | 0.1 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.0 | 0.6 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.3 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.5 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 1.7 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.1 | GO:0086002 | cardiac muscle cell action potential involved in contraction(GO:0086002) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.1 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.0 | 0.0 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.0 | 0.1 | GO:0045988 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) positive regulation of striated muscle contraction(GO:0045989) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.0 | 0.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.0 | GO:0021571 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) |
| 0.0 | 0.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.3 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.2 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.7 | GO:0050879 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 0.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.5 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.0 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.0 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.7 | GO:0072676 | lymphocyte migration(GO:0072676) |
| 0.0 | 0.4 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.4 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.5 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.1 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.0 | 0.1 | GO:0071688 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.1 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) caudal fin morphogenesis(GO:0035143) |
| 0.0 | 0.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.2 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.0 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.0 | 0.0 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.0 | 0.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.0 | GO:1903792 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.0 | 0.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.1 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.1 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.0 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.0 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.2 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.5 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 1.9 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.2 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 2.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0008352 | katanin complex(GO:0008352) |
| 0.0 | 0.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.1 | GO:0072380 | TRC complex(GO:0072380) |
| 0.0 | 2.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.3 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.1 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 2.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.9 | GO:0030055 | cell-substrate junction(GO:0030055) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.6 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.0 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.0 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 0.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 0.6 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.8 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.9 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 1.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.3 | GO:1990825 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.5 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 0.9 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.4 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.3 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 0.2 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.2 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.2 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 2.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.5 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 1.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.5 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.2 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:1990931 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.0 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.0 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.0 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.0 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 2.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.3 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |