Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
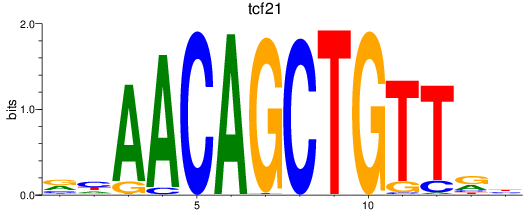
Results for tcf21
Z-value: 1.63
Transcription factors associated with tcf21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf21
|
ENSDARG00000036869 | transcription factor 21 |
|
tcf21
|
ENSDARG00000111209 | transcription factor 21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf21 | dr11_v1_chr23_-_31555696_31555696 | -0.26 | 6.8e-01 | Click! |
Activity profile of tcf21 motif
Sorted Z-values of tcf21 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_34794829 | 1.78 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr3_+_1055588 | 1.72 |
ENSDART00000143220
|
zgc:153921
|
zgc:153921 |
| chr5_+_28830643 | 1.57 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr6_+_112579 | 1.50 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr11_-_5865744 | 1.39 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr5_+_28849155 | 1.38 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr5_+_28858345 | 1.12 |
ENSDART00000111180
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr7_+_6652967 | 0.96 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr9_-_35633827 | 0.96 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr19_+_43119014 | 0.89 |
ENSDART00000023156
|
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr7_+_10610791 | 0.87 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr3_-_21094437 | 0.85 |
ENSDART00000153739
ENSDART00000109790 |
nlk1
|
nemo-like kinase, type 1 |
| chr20_-_26551210 | 0.83 |
ENSDART00000077715
|
si:dkey-25e12.3
|
si:dkey-25e12.3 |
| chr7_+_2455344 | 0.80 |
ENSDART00000172942
|
si:dkey-125e8.4
|
si:dkey-125e8.4 |
| chr6_-_8360918 | 0.80 |
ENSDART00000004716
|
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr2_+_4208323 | 0.80 |
ENSDART00000167906
|
gata6
|
GATA binding protein 6 |
| chr16_+_23972126 | 0.79 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr15_-_945804 | 0.79 |
ENSDART00000063257
|
alox5b.1
|
arachidonate 5-lipoxygenase b, tandem duplicate 1 |
| chr5_+_28857969 | 0.78 |
ENSDART00000149850
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr20_+_42668875 | 0.77 |
ENSDART00000048890
|
slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr10_-_25217347 | 0.76 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr16_-_31686602 | 0.74 |
ENSDART00000170357
|
c1s
|
complement component 1, s subcomponent |
| chr7_-_24364536 | 0.73 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr2_-_17114852 | 0.73 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr15_-_39969988 | 0.71 |
ENSDART00000146054
|
rps5
|
ribosomal protein S5 |
| chr5_+_28848870 | 0.68 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr10_+_31809226 | 0.67 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr21_-_20929575 | 0.66 |
ENSDART00000163889
|
c6
|
complement component 6 |
| chr24_+_22056386 | 0.65 |
ENSDART00000132390
|
ankrd33ba
|
ankyrin repeat domain 33ba |
| chr12_+_20693743 | 0.64 |
ENSDART00000153023
ENSDART00000153370 |
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr19_+_43119698 | 0.64 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr8_-_25247284 | 0.64 |
ENSDART00000132697
|
gnat2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr15_+_46344655 | 0.64 |
ENSDART00000155893
|
si:ch1073-340i21.2
|
si:ch1073-340i21.2 |
| chr5_+_28830388 | 0.63 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr5_-_65000312 | 0.63 |
ENSDART00000192893
|
ANXA1 (1 of many)
|
zgc:110283 |
| chr25_-_7650335 | 0.62 |
ENSDART00000089034
|
myo5ab
|
myosin VAb |
| chr8_+_45334255 | 0.62 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr14_-_34605607 | 0.61 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr20_-_35578435 | 0.60 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr11_-_183328 | 0.59 |
ENSDART00000168562
|
avil
|
advillin |
| chr8_+_39724138 | 0.59 |
ENSDART00000009323
|
pla2g1b
|
phospholipase A2, group IB (pancreas) |
| chr8_-_19280856 | 0.59 |
ENSDART00000100473
|
zgc:77486
|
zgc:77486 |
| chr10_+_35491216 | 0.59 |
ENSDART00000109705
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr12_+_30360184 | 0.58 |
ENSDART00000190718
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
| chr1_+_52560549 | 0.58 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr23_-_18057270 | 0.57 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr1_+_33390286 | 0.57 |
ENSDART00000139004
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr2_-_44344321 | 0.57 |
ENSDART00000084174
|
lig1
|
ligase I, DNA, ATP-dependent |
| chr9_-_3519253 | 0.57 |
ENSDART00000169586
|
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr23_+_29908348 | 0.57 |
ENSDART00000186622
ENSDART00000150174 |
si:ch73-236e11.2
|
si:ch73-236e11.2 |
| chr6_-_54348568 | 0.55 |
ENSDART00000156501
|
zgc:101577
|
zgc:101577 |
| chr19_+_2877079 | 0.55 |
ENSDART00000160847
|
hgh1
|
HGH1 homolog (S. cerevisiae) |
| chr22_-_16154771 | 0.55 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr5_+_25762271 | 0.54 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr7_-_8010102 | 0.54 |
ENSDART00000190305
|
si:cabz01030277.1
|
si:cabz01030277.1 |
| chr2_-_17115256 | 0.53 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr5_-_64900552 | 0.53 |
ENSDART00000073963
|
si:ch211-236k19.2
|
si:ch211-236k19.2 |
| chr10_+_29849977 | 0.53 |
ENSDART00000180242
|
hspa8
|
heat shock protein 8 |
| chr17_+_8323348 | 0.52 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr18_+_34861568 | 0.52 |
ENSDART00000192825
|
LO018333.1
|
|
| chr17_-_14836320 | 0.51 |
ENSDART00000157051
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr10_-_44560165 | 0.51 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr7_+_49664174 | 0.51 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr10_+_29850330 | 0.51 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr14_-_34605804 | 0.50 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr5_+_1911814 | 0.50 |
ENSDART00000172233
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr10_+_26667475 | 0.49 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr24_+_37709191 | 0.49 |
ENSDART00000066558
|
decr2
|
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr21_-_39253647 | 0.48 |
ENSDART00000133034
|
tusc5b
|
tumor suppressor candidate 5b |
| chr19_+_7636941 | 0.48 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr5_+_13549603 | 0.48 |
ENSDART00000171270
|
ckap2l
|
cytoskeleton associated protein 2-like |
| chr10_+_24445698 | 0.48 |
ENSDART00000146370
|
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr9_+_21146862 | 0.48 |
ENSDART00000136365
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr25_+_3326885 | 0.48 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr19_+_31541607 | 0.47 |
ENSDART00000181604
|
gmnn
|
geminin, DNA replication inhibitor |
| chr1_-_39895859 | 0.46 |
ENSDART00000135791
ENSDART00000035739 |
tmem134
|
transmembrane protein 134 |
| chr3_-_14571514 | 0.45 |
ENSDART00000137197
|
swsap1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr8_-_27687095 | 0.45 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr3_-_16537441 | 0.45 |
ENSDART00000080749
ENSDART00000133824 |
eps8l1
|
eps8-like1 |
| chr11_+_21096339 | 0.45 |
ENSDART00000124574
|
il19l
|
interleukin 19 like |
| chr7_-_6357952 | 0.45 |
ENSDART00000173197
|
zgc:165555
|
zgc:165555 |
| chr24_-_25166416 | 0.45 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr12_+_30360579 | 0.44 |
ENSDART00000152900
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
| chr17_+_8799451 | 0.44 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr5_-_29152457 | 0.44 |
ENSDART00000078469
|
noxa1
|
NADPH oxidase activator 1 |
| chr13_+_40770628 | 0.43 |
ENSDART00000085846
|
nkx1.2la
|
NK1 transcription factor related 2-like,a |
| chr24_-_25166720 | 0.43 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr9_-_49531762 | 0.43 |
ENSDART00000121875
|
xirp2b
|
xin actin binding repeat containing 2b |
| chr23_+_1730663 | 0.42 |
ENSDART00000149545
|
tgm1
|
transglutaminase 1, K polypeptide |
| chr3_+_4266289 | 0.42 |
ENSDART00000101636
|
si:dkey-73p2.1
|
si:dkey-73p2.1 |
| chr17_+_53156530 | 0.41 |
ENSDART00000126277
ENSDART00000156774 |
dph6
|
diphthamine biosynthesis 6 |
| chr11_-_35171162 | 0.41 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr6_+_56141852 | 0.41 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr5_-_24238733 | 0.40 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr19_+_9666961 | 0.40 |
ENSDART00000104596
ENSDART00000132124 |
si:dkey-14o18.6
|
si:dkey-14o18.6 |
| chr7_+_30051880 | 0.40 |
ENSDART00000075609
|
pstpip1b
|
proline-serine-threonine phosphatase interacting protein 1b |
| chr2_-_31833347 | 0.39 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr12_-_44196222 | 0.39 |
ENSDART00000171730
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr5_-_57480660 | 0.39 |
ENSDART00000147875
ENSDART00000142776 |
si:ch211-202f5.2
|
si:ch211-202f5.2 |
| chr22_+_19264181 | 0.39 |
ENSDART00000192750
ENSDART00000137591 ENSDART00000136197 |
si:dkey-78l4.5
si:dkey-21e2.11
|
si:dkey-78l4.5 si:dkey-21e2.11 |
| chr22_+_8701238 | 0.39 |
ENSDART00000106092
|
zmp:0000000652
|
zmp:0000000652 |
| chr1_-_52498146 | 0.39 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr18_+_3052408 | 0.38 |
ENSDART00000181563
|
kctd14
|
potassium channel tetramerization domain containing 14 |
| chr15_-_18197008 | 0.37 |
ENSDART00000147632
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr15_+_20403084 | 0.37 |
ENSDART00000141388
ENSDART00000152734 |
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr7_+_4971938 | 0.37 |
ENSDART00000172889
ENSDART00000145389 |
si:dkey-81n2.2
|
si:dkey-81n2.2 |
| chr2_+_36620011 | 0.37 |
ENSDART00000177428
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr13_-_37619159 | 0.37 |
ENSDART00000186348
|
zgc:152791
|
zgc:152791 |
| chr14_+_34952883 | 0.36 |
ENSDART00000182812
|
il12ba
|
interleukin 12Ba |
| chr5_+_50879545 | 0.36 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr1_-_34450784 | 0.36 |
ENSDART00000140515
|
lmo7b
|
LIM domain 7b |
| chr2_+_32016256 | 0.35 |
ENSDART00000005143
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr15_+_32727848 | 0.35 |
ENSDART00000161361
|
postnb
|
periostin, osteoblast specific factor b |
| chr16_+_41135018 | 0.35 |
ENSDART00000139435
|
si:ch211-53m15.2
|
si:ch211-53m15.2 |
| chr5_+_37068223 | 0.35 |
ENSDART00000164279
|
si:dkeyp-110c7.4
|
si:dkeyp-110c7.4 |
| chr14_+_9287683 | 0.35 |
ENSDART00000122485
|
msnb
|
moesin b |
| chr22_-_10470663 | 0.34 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr18_+_33132266 | 0.34 |
ENSDART00000151623
|
si:ch211-229c8.14
|
si:ch211-229c8.14 |
| chr13_-_24260609 | 0.34 |
ENSDART00000138747
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr8_-_32805214 | 0.34 |
ENSDART00000131597
|
zgc:194839
|
zgc:194839 |
| chr5_+_4366431 | 0.33 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr13_+_24584401 | 0.33 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr1_+_58137020 | 0.33 |
ENSDART00000180887
|
si:ch211-15j1.4
|
si:ch211-15j1.4 |
| chr18_-_7948188 | 0.32 |
ENSDART00000091805
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr7_+_34231782 | 0.32 |
ENSDART00000173547
|
lctla
|
lactase-like a |
| chr7_+_30051488 | 0.32 |
ENSDART00000169410
|
pstpip1b
|
proline-serine-threonine phosphatase interacting protein 1b |
| chr16_-_42750295 | 0.32 |
ENSDART00000176570
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr2_+_32016516 | 0.32 |
ENSDART00000135040
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr5_+_23630384 | 0.32 |
ENSDART00000013745
|
cx39.9
|
connexin 39.9 |
| chr22_+_1049367 | 0.32 |
ENSDART00000065380
|
camk1ga
|
calcium/calmodulin-dependent protein kinase IGa |
| chr14_+_80685 | 0.32 |
ENSDART00000188443
|
stag3
|
stromal antigen 3 |
| chr22_+_19230434 | 0.32 |
ENSDART00000132929
|
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr18_-_10995410 | 0.32 |
ENSDART00000136751
|
tspan33b
|
tetraspanin 33b |
| chr7_-_66864756 | 0.31 |
ENSDART00000184462
ENSDART00000189424 |
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr6_+_25261297 | 0.31 |
ENSDART00000162824
ENSDART00000163490 ENSDART00000157790 ENSDART00000160978 ENSDART00000161545 ENSDART00000159978 |
kyat3
|
kynurenine aminotransferase 3 |
| chr13_+_21601149 | 0.31 |
ENSDART00000179369
|
sh2d4ba
|
SH2 domain containing 4Ba |
| chr13_+_11876437 | 0.31 |
ENSDART00000179753
|
trim8a
|
tripartite motif containing 8a |
| chr1_+_56843411 | 0.30 |
ENSDART00000152312
|
si:ch211-152f2.1
|
si:ch211-152f2.1 |
| chr8_+_24747865 | 0.30 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr21_-_12749501 | 0.30 |
ENSDART00000179724
|
LO018011.1
|
|
| chr1_+_58228542 | 0.30 |
ENSDART00000132423
|
si:dkey-222h21.9
|
si:dkey-222h21.9 |
| chr22_-_906757 | 0.30 |
ENSDART00000193182
|
FP016205.1
|
|
| chr3_-_12227359 | 0.30 |
ENSDART00000167356
|
tfap4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr20_+_18163355 | 0.30 |
ENSDART00000011287
|
aqp4
|
aquaporin 4 |
| chr16_+_33144112 | 0.29 |
ENSDART00000183149
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr7_+_44715224 | 0.29 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr6_-_43007264 | 0.29 |
ENSDART00000124623
|
si:ch73-361p23.3
|
si:ch73-361p23.3 |
| chr8_+_22404981 | 0.29 |
ENSDART00000185211
ENSDART00000099972 |
si:dkey-23c22.7
si:dkey-23c22.9
|
si:dkey-23c22.7 si:dkey-23c22.9 |
| chr15_+_28096152 | 0.29 |
ENSDART00000100293
ENSDART00000140092 |
crybb1l3
|
crystallin, beta B1, like 3 |
| chr7_+_1473929 | 0.29 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr16_-_36905789 | 0.29 |
ENSDART00000102212
|
tdp2a
|
tyrosyl-DNA phosphodiesterase 2a |
| chr23_-_18057851 | 0.29 |
ENSDART00000173075
ENSDART00000173230 ENSDART00000173135 ENSDART00000173431 ENSDART00000173068 ENSDART00000172987 |
zgc:92287
|
zgc:92287 |
| chr11_+_44503774 | 0.28 |
ENSDART00000169295
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr14_+_21905492 | 0.28 |
ENSDART00000054437
|
tbc1d10c
|
TBC1 domain family, member 10C |
| chr1_-_40911332 | 0.28 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr5_-_19400166 | 0.28 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr6_+_58915889 | 0.28 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr15_+_6459847 | 0.28 |
ENSDART00000157250
ENSDART00000065824 |
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr7_-_24046999 | 0.28 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr1_-_52497834 | 0.28 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr2_-_51741711 | 0.27 |
ENSDART00000171301
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr1_-_34450622 | 0.27 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr3_-_34724879 | 0.27 |
ENSDART00000177021
|
thraa
|
thyroid hormone receptor alpha a |
| chr12_-_9294819 | 0.27 |
ENSDART00000003805
|
pth1rb
|
parathyroid hormone 1 receptor b |
| chr4_-_43374443 | 0.27 |
ENSDART00000191022
|
BX927253.3
|
|
| chr11_-_35171768 | 0.27 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr12_+_2660733 | 0.26 |
ENSDART00000152235
|
irbpl
|
interphotoreceptor retinoid-binding protein like |
| chr18_+_2228737 | 0.26 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr11_+_29975830 | 0.26 |
ENSDART00000148929
|
si:ch73-226l13.2
|
si:ch73-226l13.2 |
| chr6_+_42475730 | 0.26 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr4_+_7508316 | 0.26 |
ENSDART00000170924
ENSDART00000170933 ENSDART00000164985 ENSDART00000167571 ENSDART00000158843 ENSDART00000158999 |
tnnt2e
|
troponin T2e, cardiac |
| chr9_+_3519191 | 0.26 |
ENSDART00000008606
|
mettl8
|
methyltransferase like 8 |
| chr15_+_37973197 | 0.25 |
ENSDART00000156661
|
si:dkey-238d18.9
|
si:dkey-238d18.9 |
| chr14_+_32837914 | 0.25 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr3_+_14571813 | 0.25 |
ENSDART00000146728
ENSDART00000171731 |
znf653
|
zinc finger protein 653 |
| chr16_-_22004402 | 0.24 |
ENSDART00000145100
ENSDART00000188217 ENSDART00000180083 |
si:dkey-71b5.7
|
si:dkey-71b5.7 |
| chr8_+_22405477 | 0.24 |
ENSDART00000148267
|
si:dkey-23c22.7
|
si:dkey-23c22.7 |
| chr2_+_44977889 | 0.24 |
ENSDART00000144024
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr19_-_10432134 | 0.24 |
ENSDART00000081440
|
il11b
|
interleukin 11b |
| chr4_+_76353418 | 0.24 |
ENSDART00000171289
|
si:ch73-158p21.2
|
si:ch73-158p21.2 |
| chr6_-_39344259 | 0.23 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr9_-_12811936 | 0.23 |
ENSDART00000188490
|
myo10l3
|
myosin X-like 3 |
| chr15_-_38009344 | 0.23 |
ENSDART00000157094
|
si:dkey-238d18.6
|
si:dkey-238d18.6 |
| chr21_-_293146 | 0.23 |
ENSDART00000157781
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr23_-_19831739 | 0.23 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr5_-_23729590 | 0.23 |
ENSDART00000014831
|
si:dkey-110k5.10
|
si:dkey-110k5.10 |
| chr12_-_33859950 | 0.23 |
ENSDART00000131181
|
sema4gb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Gb |
| chr5_+_64900223 | 0.22 |
ENSDART00000191677
|
ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr15_-_25365319 | 0.22 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr2_+_17508323 | 0.22 |
ENSDART00000169703
|
pimr199
|
Pim proto-oncogene, serine/threonine kinase, related 199 |
| chr10_+_1590604 | 0.22 |
ENSDART00000166646
|
ora5
|
olfactory receptor class A related 5 |
| chr21_+_19418563 | 0.22 |
ENSDART00000181113
ENSDART00000080110 |
amacr
|
alpha-methylacyl-CoA racemase |
| chr10_-_20065149 | 0.21 |
ENSDART00000182603
|
BX323022.1
|
|
| chr3_+_58379450 | 0.21 |
ENSDART00000155759
|
sdr42e2
|
short chain dehydrogenase/reductase family 42E, member 2 |
| chr21_-_37435162 | 0.21 |
ENSDART00000133585
|
fam114a2
|
family with sequence similarity 114, member A2 |
| chr19_+_23308419 | 0.21 |
ENSDART00000141558
|
irgf2
|
immunity-related GTPase family, f2 |
| chr2_-_7131657 | 0.21 |
ENSDART00000175565
ENSDART00000092116 |
extl2
|
exostosin-like glycosyltransferase 2 |
| chr4_-_76154252 | 0.21 |
ENSDART00000161269
|
BX957252.1
|
|
| chr23_-_44470253 | 0.21 |
ENSDART00000176333
|
si:ch1073-228j22.2
|
si:ch1073-228j22.2 |
| chr2_-_49997055 | 0.20 |
ENSDART00000140294
|
si:ch211-106n13.3
|
si:ch211-106n13.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf21
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.3 | 0.8 | GO:0050995 | negative regulation of lipid transport(GO:0032369) negative regulation of lipid catabolic process(GO:0050995) |
| 0.3 | 0.8 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.2 | 1.0 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.2 | 0.8 | GO:0043011 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.6 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.9 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.4 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 1.3 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 0.6 | GO:0006266 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.6 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.1 | 0.7 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 0.5 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.3 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.6 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.2 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.1 | 0.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.8 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 6.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.4 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.8 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.4 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.3 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.6 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.5 | GO:2000403 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.6 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.3 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.4 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.3 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.4 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0015860 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.3 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.2 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.5 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 1.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.3 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 0.7 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.5 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.3 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.0 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 0.4 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.1 | 1.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.8 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.1 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.0 | 0.5 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.0 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.3 | GO:0045178 | basal part of cell(GO:0045178) basal cortex(GO:0045180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 0.6 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 0.9 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 0.5 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 0.6 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.8 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 0.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 1.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.3 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.4 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.1 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 1.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.8 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 0.3 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.8 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.5 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 5.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.0 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.1 | GO:0015217 | ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 1.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.6 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0005501 | retinoid binding(GO:0005501) isoprenoid binding(GO:0019840) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 2.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.6 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.2 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 1.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |