Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
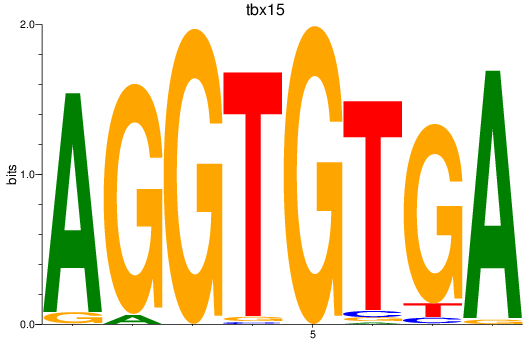
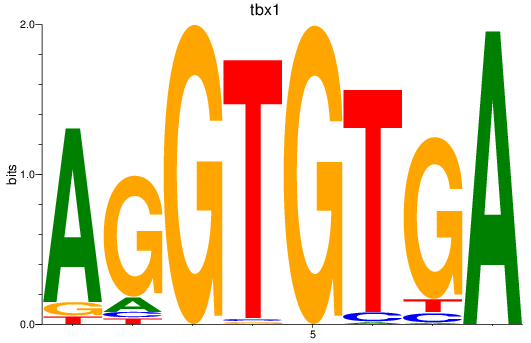
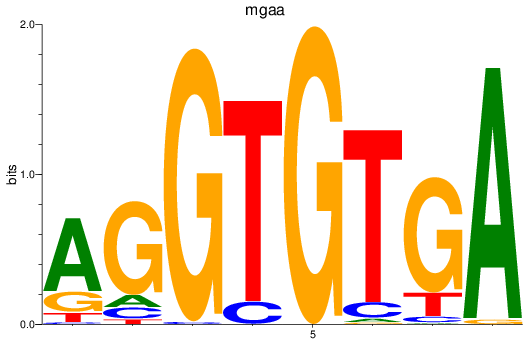
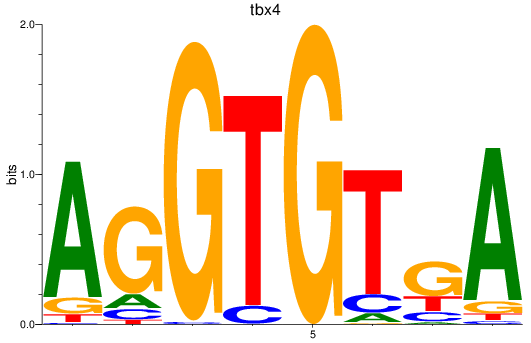
Results for tbx15_tbx1_mgaa_tbx4
Z-value: 2.05
Transcription factors associated with tbx15_tbx1_mgaa_tbx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx15
|
ENSDARG00000002582 | T-box transcription factor 15 |
|
tbx1
|
ENSDARG00000031891 | T-box transcription factor 1 |
|
mgaa
|
ENSDARG00000078784 | MAX dimerization protein MGA a |
|
tbx4
|
ENSDARG00000030058 | T-box transcription factor 4 |
|
tbx4
|
ENSDARG00000113067 | T-box transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mgaa | dr11_v1_chr17_+_10566490_10566495 | 0.93 | 2.2e-02 | Click! |
| tbx15 | dr11_v1_chr9_-_21067673_21067673 | 0.59 | 2.9e-01 | Click! |
| tbx4 | dr11_v1_chr15_+_27384798_27384798 | 0.44 | 4.6e-01 | Click! |
| tbx1 | dr11_v1_chr5_+_15203421_15203421 | -0.39 | 5.2e-01 | Click! |
Activity profile of tbx15_tbx1_mgaa_tbx4 motif
Sorted Z-values of tbx15_tbx1_mgaa_tbx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_37837245 | 4.95 |
ENSDART00000171617
|
epd
|
ependymin |
| chr25_+_37397031 | 3.28 |
ENSDART00000193643
ENSDART00000169132 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr7_-_38612230 | 2.69 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr24_-_6158933 | 2.61 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr23_-_4855122 | 2.59 |
ENSDART00000133701
|
slc6a1a
|
solute carrier family 6 (neurotransmitter transporter), member 1a |
| chr10_-_31782616 | 2.09 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr21_-_43550120 | 1.86 |
ENSDART00000151627
|
si:ch73-362m14.2
|
si:ch73-362m14.2 |
| chr19_+_14921000 | 1.75 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr25_+_7784582 | 1.72 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr22_-_20575679 | 1.70 |
ENSDART00000089033
|
lingo3a
|
leucine rich repeat and Ig domain containing 3a |
| chr16_-_12173399 | 1.66 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr5_-_33215261 | 1.64 |
ENSDART00000097935
ENSDART00000134777 |
si:dkey-226m8.10
|
si:dkey-226m8.10 |
| chr7_+_7048245 | 1.62 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr3_-_61205711 | 1.59 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr13_-_27653679 | 1.59 |
ENSDART00000142568
|
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr5_-_46896541 | 1.58 |
ENSDART00000133240
|
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr16_+_30301539 | 1.51 |
ENSDART00000186018
|
LO017848.1
|
|
| chr16_-_12173554 | 1.50 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr7_+_30867008 | 1.50 |
ENSDART00000193106
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr16_-_13109749 | 1.49 |
ENSDART00000142610
|
prkcg
|
protein kinase C, gamma |
| chr16_-_29437373 | 1.45 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr7_+_6969909 | 1.38 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr18_-_20822045 | 1.38 |
ENSDART00000100743
|
cttnbp2
|
cortactin binding protein 2 |
| chr9_+_34334156 | 1.36 |
ENSDART00000144272
|
pou2f1b
|
POU class 2 homeobox 1b |
| chr21_-_43949208 | 1.35 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr23_+_28582865 | 1.34 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr12_-_35386910 | 1.32 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr16_-_17207754 | 1.31 |
ENSDART00000063804
|
wu:fj39g12
|
wu:fj39g12 |
| chr15_-_16098531 | 1.28 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr17_+_27176243 | 1.21 |
ENSDART00000162527
|
si:ch211-160f23.7
|
si:ch211-160f23.7 |
| chr2_-_32501501 | 1.19 |
ENSDART00000181309
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr14_-_26177156 | 1.18 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr11_+_25583950 | 1.17 |
ENSDART00000111961
|
ccdc120
|
coiled-coil domain containing 120 |
| chr18_+_22302635 | 1.14 |
ENSDART00000141051
|
carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr5_+_22791686 | 1.11 |
ENSDART00000014806
|
npas2
|
neuronal PAS domain protein 2 |
| chr11_+_23933016 | 1.11 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr13_+_4205724 | 1.10 |
ENSDART00000134105
|
dlk2
|
delta-like 2 homolog (Drosophila) |
| chr22_+_11857356 | 1.08 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr10_-_31562695 | 1.07 |
ENSDART00000186456
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr21_+_44300689 | 1.02 |
ENSDART00000186298
ENSDART00000142810 |
gabra3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr1_+_8662530 | 1.01 |
ENSDART00000054989
|
fscn1b
|
fascin actin-bundling protein 1b |
| chr19_+_47311869 | 1.01 |
ENSDART00000136647
|
ext1c
|
exostoses (multiple) 1c |
| chr16_+_50741154 | 1.00 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr3_-_33901483 | 0.98 |
ENSDART00000144774
ENSDART00000138765 |
cacna1aa
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, a |
| chr6_+_40587551 | 0.96 |
ENSDART00000155554
|
frs3
|
fibroblast growth factor receptor substrate 3 |
| chr22_-_14475927 | 0.90 |
ENSDART00000135768
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr17_+_28340138 | 0.90 |
ENSDART00000033943
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr21_-_10488379 | 0.89 |
ENSDART00000163878
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr23_-_30431333 | 0.88 |
ENSDART00000146633
|
camta1a
|
calmodulin binding transcription activator 1a |
| chr7_-_51476276 | 0.88 |
ENSDART00000082464
|
nhsl2
|
NHS-like 2 |
| chr22_-_12160283 | 0.86 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr15_-_18432673 | 0.85 |
ENSDART00000146853
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr5_+_59449762 | 0.85 |
ENSDART00000150230
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr21_-_39628771 | 0.84 |
ENSDART00000183995
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr18_-_51015718 | 0.83 |
ENSDART00000190698
|
LO018598.1
|
|
| chr1_-_23110740 | 0.83 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr1_-_8101495 | 0.81 |
ENSDART00000161938
|
si:dkeyp-9d4.3
|
si:dkeyp-9d4.3 |
| chr6_+_13606410 | 0.81 |
ENSDART00000104716
|
asic4b
|
acid-sensing (proton-gated) ion channel family member 4b |
| chr13_-_36264861 | 0.80 |
ENSDART00000100204
|
slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr3_-_33469722 | 0.79 |
ENSDART00000151539
|
sgsm3
|
small G protein signaling modulator 3 |
| chr20_-_15922210 | 0.79 |
ENSDART00000152412
ENSDART00000152354 ENSDART00000152828 ENSDART00000013453 ENSDART00000152357 |
fam20b
|
family with sequence similarity 20, member B (H. sapiens) |
| chr16_+_5702749 | 0.79 |
ENSDART00000138330
|
zgc:158689
|
zgc:158689 |
| chr9_+_32978302 | 0.77 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr20_+_41549200 | 0.77 |
ENSDART00000135715
|
fam184a
|
family with sequence similarity 184, member A |
| chr19_-_5812319 | 0.76 |
ENSDART00000114472
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr7_+_46368520 | 0.76 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr13_-_21739142 | 0.75 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr13_-_24880525 | 0.75 |
ENSDART00000136624
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr12_-_26383242 | 0.75 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr2_+_33368414 | 0.74 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr7_-_55675617 | 0.74 |
ENSDART00000021009
ENSDART00000188631 |
cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr11_+_1628538 | 0.73 |
ENSDART00000154967
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr1_-_40341306 | 0.73 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr5_+_32228538 | 0.72 |
ENSDART00000077471
|
myhc4
|
myosin heavy chain 4 |
| chr23_+_40604951 | 0.71 |
ENSDART00000114959
|
cdh24a
|
cadherin 24, type 2a |
| chr22_-_15385442 | 0.70 |
ENSDART00000090975
|
tmem264
|
transmembrane protein 264 |
| chr14_+_6605675 | 0.70 |
ENSDART00000143179
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr19_+_32947910 | 0.69 |
ENSDART00000052091
|
atp6v1c1b
|
ATPase H+ transporting V1 subunit C1b |
| chr13_-_30027730 | 0.69 |
ENSDART00000044009
|
scdb
|
stearoyl-CoA desaturase b |
| chr4_-_4795205 | 0.69 |
ENSDART00000039313
|
zgc:162331
|
zgc:162331 |
| chr16_+_53203370 | 0.69 |
ENSDART00000154669
|
si:ch211-269k10.2
|
si:ch211-269k10.2 |
| chr9_-_41323746 | 0.68 |
ENSDART00000140564
|
glsb
|
glutaminase b |
| chr4_-_19016396 | 0.67 |
ENSDART00000166160
|
si:dkey-31f5.11
|
si:dkey-31f5.11 |
| chr19_-_28360033 | 0.67 |
ENSDART00000186994
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr13_-_24906307 | 0.66 |
ENSDART00000148191
ENSDART00000189810 |
kat6b
|
K(lysine) acetyltransferase 6B |
| chr17_-_20394566 | 0.66 |
ENSDART00000154667
|
sorcs3b
|
sortilin related VPS10 domain containing receptor 3b |
| chr20_+_41549735 | 0.66 |
ENSDART00000184235
|
fam184a
|
family with sequence similarity 184, member A |
| chr23_+_45584223 | 0.66 |
ENSDART00000149367
|
si:ch73-290k24.5
|
si:ch73-290k24.5 |
| chr18_-_13360106 | 0.65 |
ENSDART00000091512
|
cmip
|
c-Maf inducing protein |
| chr4_+_11384891 | 0.65 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr3_-_25814097 | 0.63 |
ENSDART00000169706
|
ntn1b
|
netrin 1b |
| chr12_-_5448993 | 0.63 |
ENSDART00000181802
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr14_+_50770537 | 0.63 |
ENSDART00000158723
|
sncb
|
synuclein, beta |
| chr12_+_32292564 | 0.61 |
ENSDART00000152945
|
ANKFN1
|
si:ch211-277e21.2 |
| chr18_+_19419120 | 0.61 |
ENSDART00000025107
|
map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr20_+_21268795 | 0.61 |
ENSDART00000090016
|
nudt14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
| chr21_+_26726936 | 0.61 |
ENSDART00000065392
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr5_-_45894802 | 0.59 |
ENSDART00000097648
|
crfb6
|
cytokine receptor family member b6 |
| chr22_-_34872533 | 0.59 |
ENSDART00000167176
|
slit1b
|
slit homolog 1b (Drosophila) |
| chr4_-_60049792 | 0.59 |
ENSDART00000158199
|
znf1033
|
zinc finger protein 1033 |
| chr17_-_3986236 | 0.59 |
ENSDART00000188794
ENSDART00000160830 |
PLCB1
|
si:ch1073-140o9.2 |
| chr21_-_25685422 | 0.58 |
ENSDART00000182921
|
phkg1b
|
phosphorylase kinase, gamma 1b (muscle) |
| chr9_+_42095220 | 0.58 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr9_+_17438765 | 0.58 |
ENSDART00000138953
|
rgcc
|
regulator of cell cycle |
| chr13_+_11439486 | 0.57 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr8_+_47571211 | 0.57 |
ENSDART00000131460
|
plch2a
|
phospholipase C, eta 2a |
| chr10_+_18952271 | 0.57 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr11_+_41540862 | 0.57 |
ENSDART00000173210
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr12_-_17147473 | 0.57 |
ENSDART00000106035
|
stambpl1
|
STAM binding protein-like 1 |
| chr14_+_45406299 | 0.56 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr19_+_22727940 | 0.56 |
ENSDART00000052509
|
trhrb
|
thyrotropin-releasing hormone receptor b |
| chr19_+_14059349 | 0.55 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr7_-_32599669 | 0.55 |
ENSDART00000173752
|
kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr3_-_42016693 | 0.55 |
ENSDART00000184741
|
ttyh3a
|
tweety family member 3a |
| chr3_+_33367954 | 0.55 |
ENSDART00000103161
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr1_-_53750522 | 0.54 |
ENSDART00000190755
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr8_+_13364950 | 0.54 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr15_+_28482862 | 0.54 |
ENSDART00000015286
ENSDART00000154320 |
ankrd13b
|
ankyrin repeat domain 13B |
| chr5_+_28398449 | 0.53 |
ENSDART00000165292
|
nsmfb
|
NMDA receptor synaptonuclear signaling and neuronal migration factor b |
| chr20_-_28800999 | 0.53 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr10_+_37500234 | 0.52 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr14_+_50937757 | 0.52 |
ENSDART00000163865
|
rnf44
|
ring finger protein 44 |
| chr5_+_42467867 | 0.52 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr2_+_23823622 | 0.52 |
ENSDART00000099581
|
si:dkey-24c2.9
|
si:dkey-24c2.9 |
| chr1_-_714626 | 0.51 |
ENSDART00000161072
|
adamts5
|
ADAM metallopeptidase with thrombospondin type 1 motif 5 |
| chr4_+_11375894 | 0.50 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr14_+_45925810 | 0.50 |
ENSDART00000189543
|
flrt1b
|
fibronectin leucine rich transmembrane protein 1b |
| chr3_-_25813426 | 0.50 |
ENSDART00000039482
|
ntn1b
|
netrin 1b |
| chr1_+_37195465 | 0.50 |
ENSDART00000043855
ENSDART00000192580 ENSDART00000181666 |
dclk2a
|
doublecortin-like kinase 2a |
| chr19_+_37458610 | 0.49 |
ENSDART00000103151
|
dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr11_-_21303946 | 0.48 |
ENSDART00000185786
|
RASSF5
|
si:dkey-85p17.3 |
| chr25_-_12788370 | 0.48 |
ENSDART00000158551
|
slc7a5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr17_-_26537928 | 0.48 |
ENSDART00000155692
ENSDART00000122366 |
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr11_-_26832685 | 0.47 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr23_+_24611747 | 0.47 |
ENSDART00000134978
|
nckap5l
|
NCK-associated protein 5-like |
| chr14_-_33872092 | 0.47 |
ENSDART00000111903
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr15_+_42397125 | 0.47 |
ENSDART00000169751
|
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr8_-_1838315 | 0.46 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr24_+_39158283 | 0.46 |
ENSDART00000053139
|
atp6v0cb
|
ATPase H+ transporting V0 subunit cb |
| chr14_-_31059218 | 0.45 |
ENSDART00000111691
ENSDART00000021379 ENSDART00000113479 |
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr1_-_17663108 | 0.45 |
ENSDART00000131559
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr5_-_37875636 | 0.44 |
ENSDART00000184674
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr18_+_45796096 | 0.43 |
ENSDART00000087070
|
abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr2_-_37893910 | 0.43 |
ENSDART00000143027
|
hbl2
|
hexose-binding lectin 2 |
| chr17_-_14726824 | 0.43 |
ENSDART00000162947
|
si:ch73-305o9.3
|
si:ch73-305o9.3 |
| chr21_+_33503835 | 0.43 |
ENSDART00000125658
|
clint1b
|
clathrin interactor 1b |
| chr24_-_36095526 | 0.42 |
ENSDART00000158145
|
CABZ01075509.1
|
|
| chr5_+_26075230 | 0.42 |
ENSDART00000098473
|
klf9
|
Kruppel-like factor 9 |
| chr1_-_30039331 | 0.42 |
ENSDART00000086935
ENSDART00000143800 |
MARCH4 (1 of many)
|
zgc:153256 |
| chr11_-_37880492 | 0.42 |
ENSDART00000102868
|
etnk2
|
ethanolamine kinase 2 |
| chr9_-_21970067 | 0.42 |
ENSDART00000009920
|
lmo7a
|
LIM domain 7a |
| chr13_-_39830816 | 0.42 |
ENSDART00000138981
|
zgc:171482
|
zgc:171482 |
| chr16_-_13613475 | 0.41 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr21_-_2217685 | 0.41 |
ENSDART00000159315
|
si:dkey-50i6.5
|
si:dkey-50i6.5 |
| chr6_-_16394528 | 0.41 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr14_-_32744464 | 0.41 |
ENSDART00000075617
|
sox3
|
SRY (sex determining region Y)-box 3 |
| chr13_-_33822550 | 0.40 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr8_-_19246342 | 0.40 |
ENSDART00000147172
|
abhd17ab
|
abhydrolase domain containing 17Ab |
| chr8_+_54055390 | 0.40 |
ENSDART00000102696
|
magi1a
|
membrane associated guanylate kinase, WW and PDZ domain containing 1a |
| chr7_-_18656069 | 0.40 |
ENSDART00000021559
|
coro1b
|
coronin, actin binding protein, 1B |
| chr9_-_18877597 | 0.40 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr10_+_43994471 | 0.39 |
ENSDART00000138242
ENSDART00000186359 |
cldn5b
|
claudin 5b |
| chr19_+_13375838 | 0.39 |
ENSDART00000163093
|
lrp12
|
low density lipoprotein receptor-related protein 12 |
| chr7_+_11197022 | 0.39 |
ENSDART00000185236
|
cemip
|
cell migration inducing protein, hyaluronan binding |
| chr15_+_40665310 | 0.39 |
ENSDART00000154187
ENSDART00000042082 |
fat3a
|
FAT atypical cadherin 3a |
| chr3_+_23488652 | 0.39 |
ENSDART00000126282
|
nr1d1
|
nuclear receptor subfamily 1, group d, member 1 |
| chr13_+_2894536 | 0.39 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr2_-_16224083 | 0.39 |
ENSDART00000165953
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr11_-_12998400 | 0.38 |
ENSDART00000018614
|
chrna4b
|
cholinergic receptor, nicotinic, alpha 4b |
| chr2_+_24203229 | 0.38 |
ENSDART00000138088
|
map4l
|
microtubule associated protein 4 like |
| chr14_-_33872616 | 0.38 |
ENSDART00000162840
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr2_+_24199276 | 0.38 |
ENSDART00000140575
|
map4l
|
microtubule associated protein 4 like |
| chr10_-_20588787 | 0.38 |
ENSDART00000138045
ENSDART00000181885 ENSDART00000091115 |
nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr22_-_21755524 | 0.38 |
ENSDART00000149635
|
tle2b
|
transducin like enhancer of split 2b |
| chr5_+_62052538 | 0.38 |
ENSDART00000141574
|
si:dkey-35m8.1
|
si:dkey-35m8.1 |
| chr8_+_1651821 | 0.37 |
ENSDART00000060865
ENSDART00000186304 |
rasal1b
|
RAS protein activator like 1b (GAP1 like) |
| chr13_+_30054996 | 0.37 |
ENSDART00000110061
ENSDART00000186045 |
spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr8_-_21268303 | 0.37 |
ENSDART00000067211
|
gpr37l1b
|
G protein-coupled receptor 37 like 1b |
| chr6_+_58406014 | 0.37 |
ENSDART00000044241
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr23_+_27912079 | 0.37 |
ENSDART00000171859
|
BX539336.1
|
|
| chr18_+_6536293 | 0.37 |
ENSDART00000024576
|
fkbp4
|
FK506 binding protein 4 |
| chr22_+_1300587 | 0.37 |
ENSDART00000124161
|
si:ch73-138e16.5
|
si:ch73-138e16.5 |
| chr8_+_8532407 | 0.37 |
ENSDART00000169276
ENSDART00000138993 |
grm6a
|
glutamate receptor, metabotropic 6a |
| chr24_+_26039464 | 0.37 |
ENSDART00000131017
|
tnk2a
|
tyrosine kinase, non-receptor, 2a |
| chr14_+_35691889 | 0.36 |
ENSDART00000074685
|
glrbb
|
glycine receptor, beta b |
| chr24_+_14713776 | 0.36 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr16_+_50100420 | 0.36 |
ENSDART00000128167
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr14_+_790166 | 0.36 |
ENSDART00000123912
|
adra2da
|
adrenergic, alpha-2D-, receptor a |
| chr10_+_42462577 | 0.36 |
ENSDART00000133463
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr17_+_9009098 | 0.36 |
ENSDART00000180856
|
akap6
|
A kinase (PRKA) anchor protein 6 |
| chr14_-_48187742 | 0.36 |
ENSDART00000129871
|
rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr3_+_16449490 | 0.36 |
ENSDART00000141467
|
kcnj12b
|
potassium inwardly-rectifying channel, subfamily J, member 12b |
| chr13_+_18276187 | 0.35 |
ENSDART00000141009
|
tet1
|
tet methylcytosine dioxygenase 1 |
| chr18_-_893574 | 0.35 |
ENSDART00000150959
|
parp6a
|
poly (ADP-ribose) polymerase family, member 6a |
| chr10_-_26430168 | 0.35 |
ENSDART00000128894
|
dchs1b
|
dachsous cadherin-related 1b |
| chr15_-_21669618 | 0.35 |
ENSDART00000156995
|
sorl1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr17_+_58211 | 0.35 |
ENSDART00000157642
|
si:ch1073-209e23.1
|
si:ch1073-209e23.1 |
| chr1_-_51606552 | 0.35 |
ENSDART00000130828
|
cnrip1a
|
cannabinoid receptor interacting protein 1a |
| chr10_-_8032885 | 0.34 |
ENSDART00000188619
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr11_+_30161168 | 0.34 |
ENSDART00000157385
|
cdkl5
|
cyclin-dependent kinase-like 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx15_tbx1_mgaa_tbx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.5 | 1.8 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.4 | 1.7 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.4 | 3.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.4 | 1.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.4 | 1.1 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.3 | 0.9 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.2 | 0.7 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.2 | 0.7 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 0.5 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.2 | 4.0 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 0.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.7 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.1 | 0.4 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.4 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.1 | 1.1 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 2.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.5 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.3 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.8 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.3 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.1 | 0.8 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.3 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 1.3 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 0.7 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.9 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 1.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.2 | GO:0098725 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.1 | 1.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.4 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 0.4 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 0.3 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 0.3 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.2 | GO:0009258 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.6 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 0.2 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.1 | 0.2 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 1.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.0 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 0.4 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.3 | GO:0048714 | positive regulation of gliogenesis(GO:0014015) positive regulation of glial cell differentiation(GO:0045687) regulation of oligodendrocyte differentiation(GO:0048713) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.4 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.6 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.3 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.4 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 4.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.1 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 1.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 1.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.9 | GO:0021602 | cranial nerve morphogenesis(GO:0021602) |
| 0.0 | 1.3 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.6 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 1.9 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.3 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.4 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.4 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.5 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 1.8 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.3 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 1.6 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.1 | GO:0002834 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.8 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 1.6 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 3.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.3 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.3 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.2 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0048521 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.2 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 1.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.5 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0060031 | mediolateral intercalation(GO:0060031) |
| 0.0 | 0.2 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.6 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 1.8 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.2 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.0 | 0.1 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.0 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.8 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.3 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 1.6 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 1.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.4 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.3 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.2 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 0.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.0 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.2 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0031063 | regulation of histone deacetylation(GO:0031063) regulation of protein deacetylation(GO:0090311) |
| 0.0 | 0.1 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.0 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.9 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.5 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.1 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.2 | 1.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.1 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 1.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.7 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 1.6 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.1 | 0.3 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 0.8 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.2 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.8 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.0 | GO:0032589 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 2.9 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.1 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.6 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 9.0 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 6.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 1.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 1.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 8.1 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.6 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.6 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.9 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.6 | 1.7 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.3 | 1.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 2.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 1.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 0.6 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.2 | 2.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 0.7 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.6 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.1 | 0.7 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 1.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.2 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.1 | 0.7 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 3.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.8 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.1 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.3 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.4 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.7 | GO:0030955 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.1 | 1.6 | GO:0009931 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.1 | 1.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.4 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.4 | GO:0016933 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 2.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0032034 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.5 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.0 | 0.2 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 4.0 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 19.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0043121 | neurotrophin binding(GO:0043121) |
| 0.0 | 0.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.3 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.2 | GO:0004952 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 3.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 3.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 1.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 2.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.9 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.5 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.2 | 2.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 0.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.0 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 1.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |