Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
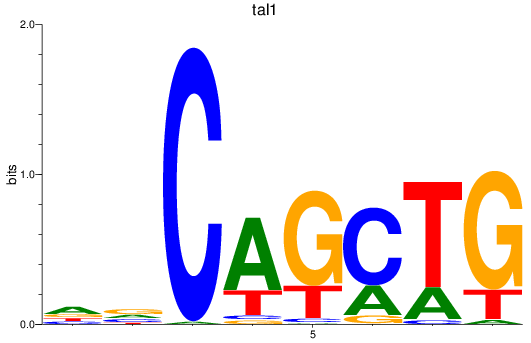
Results for tal1
Z-value: 2.11
Transcription factors associated with tal1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tal1
|
ENSDARG00000019930 | T-cell acute lymphocytic leukemia 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tal1 | dr11_v1_chr22_+_16535575_16535575 | -0.42 | 4.8e-01 | Click! |
Activity profile of tal1 motif
Sorted Z-values of tal1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_19790962 | 1.83 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr25_-_18470695 | 1.77 |
ENSDART00000034377
|
cpa5
|
carboxypeptidase A5 |
| chr3_-_3209432 | 1.59 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr10_-_342564 | 1.58 |
ENSDART00000157633
|
vaspa
|
vasodilator stimulated phosphoprotein a |
| chr3_+_58833306 | 1.55 |
ENSDART00000113223
|
igl1c3
|
immunoglobulin light 1 constant 3 |
| chr16_-_41990421 | 1.38 |
ENSDART00000055921
|
pycard
|
PYD and CARD domain containing |
| chr22_-_10541372 | 1.38 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr21_+_5531138 | 1.31 |
ENSDART00000163825
|
ly6m6
|
lymphocyte antigen 6 family member M6 |
| chr13_+_669177 | 1.19 |
ENSDART00000190085
|
CU462915.1
|
|
| chr7_+_71535045 | 1.15 |
ENSDART00000047069
|
tyms
|
thymidylate synthetase |
| chr3_-_61181018 | 1.15 |
ENSDART00000187970
|
pvalb4
|
parvalbumin 4 |
| chr19_+_48117995 | 1.14 |
ENSDART00000170865
|
nme2b.1
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 |
| chr23_+_44614056 | 1.12 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr12_+_13256415 | 1.11 |
ENSDART00000144542
|
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr7_+_39389273 | 1.06 |
ENSDART00000191298
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr6_+_40629066 | 1.04 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr5_-_60885935 | 1.03 |
ENSDART00000128350
|
rad51d
|
RAD51 paralog D |
| chr3_+_3598555 | 1.02 |
ENSDART00000191152
|
CR589947.3
|
|
| chr3_+_4266289 | 1.00 |
ENSDART00000101636
|
si:dkey-73p2.1
|
si:dkey-73p2.1 |
| chr2_+_45191049 | 0.99 |
ENSDART00000165392
|
ccl20a.3
|
chemokine (C-C motif) ligand 20a, duplicate 3 |
| chr7_+_8456999 | 0.99 |
ENSDART00000172880
|
jac4
|
jacalin 4 |
| chr4_+_77943184 | 0.98 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr5_+_338154 | 0.97 |
ENSDART00000191743
|
rnf170
|
ring finger protein 170 |
| chr21_+_10577527 | 0.97 |
ENSDART00000165070
ENSDART00000127963 |
ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr22_+_20720808 | 0.96 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr19_-_25519612 | 0.95 |
ENSDART00000133150
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr2_-_7666021 | 0.95 |
ENSDART00000180007
|
CABZ01021592.1
|
|
| chr1_-_59141715 | 0.95 |
ENSDART00000164941
ENSDART00000138870 |
si:ch1073-110a20.1
|
si:ch1073-110a20.1 |
| chr7_-_8438657 | 0.94 |
ENSDART00000173054
|
si:dkeyp-32g11.8
|
si:dkeyp-32g11.8 |
| chr13_-_438705 | 0.93 |
ENSDART00000082142
|
CU570800.1
|
|
| chr9_+_307863 | 0.93 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr20_+_2281933 | 0.92 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr17_-_2584423 | 0.90 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr4_+_38550788 | 0.90 |
ENSDART00000157412
|
si:ch211-209n20.1
|
si:ch211-209n20.1 |
| chr24_-_6029314 | 0.87 |
ENSDART00000136155
|
ftr60
|
finTRIM family, member 60 |
| chr11_+_6136220 | 0.85 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr21_+_5974590 | 0.84 |
ENSDART00000098499
|
dck
|
deoxycytidine kinase |
| chr3_-_3939785 | 0.84 |
ENSDART00000049593
|
unm_sa1506
|
un-named sa1506 |
| chr23_+_16807945 | 0.83 |
ENSDART00000080660
|
zgc:114081
|
zgc:114081 |
| chr2_+_11031360 | 0.82 |
ENSDART00000180020
ENSDART00000145093 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr25_-_18454016 | 0.81 |
ENSDART00000005877
|
cpa1
|
carboxypeptidase A1 (pancreatic) |
| chr20_+_172162 | 0.81 |
ENSDART00000168765
|
BX511077.1
|
|
| chr9_+_33220342 | 0.81 |
ENSDART00000100893
ENSDART00000113451 |
si:ch211-125e6.13
|
si:ch211-125e6.13 |
| chr21_-_217589 | 0.80 |
ENSDART00000185017
|
CZQB01146713.1
|
|
| chr23_+_16807785 | 0.79 |
ENSDART00000146992
|
zgc:114081
|
zgc:114081 |
| chr22_-_26353916 | 0.78 |
ENSDART00000077958
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr4_-_16354292 | 0.78 |
ENSDART00000139919
|
lum
|
lumican |
| chr15_+_47525073 | 0.77 |
ENSDART00000067583
|
sidt2
|
SID1 transmembrane family, member 2 |
| chr3_-_4574372 | 0.77 |
ENSDART00000172023
ENSDART00000168724 |
ftr50
|
finTRIM family, member 50 |
| chr3_-_47235997 | 0.76 |
ENSDART00000047071
|
tmed1a
|
transmembrane p24 trafficking protein 1a |
| chr7_+_2455344 | 0.76 |
ENSDART00000172942
|
si:dkey-125e8.4
|
si:dkey-125e8.4 |
| chr1_-_52494122 | 0.75 |
ENSDART00000131407
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr9_+_51225345 | 0.75 |
ENSDART00000132896
|
fap
|
fibroblast activation protein, alpha |
| chr8_-_27687095 | 0.75 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr2_-_45191319 | 0.74 |
ENSDART00000192272
|
CR407590.2
|
|
| chr25_+_15933411 | 0.73 |
ENSDART00000191581
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr3_-_44012748 | 0.72 |
ENSDART00000167248
ENSDART00000157463 ENSDART00000159111 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr18_+_38321039 | 0.72 |
ENSDART00000132534
ENSDART00000111260 ENSDART00000192806 |
alx4b
|
ALX homeobox 4b |
| chr7_+_56577522 | 0.72 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr3_+_3545825 | 0.72 |
ENSDART00000109060
|
CR589947.1
|
|
| chr12_-_48168135 | 0.72 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr17_-_2595736 | 0.72 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr19_-_48010490 | 0.70 |
ENSDART00000159938
|
FBXL19
|
zgc:158376 |
| chr7_+_35229805 | 0.70 |
ENSDART00000173911
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr22_+_987788 | 0.70 |
ENSDART00000149486
|
def6b
|
differentially expressed in FDCP 6b homolog (mouse) |
| chr3_+_54581987 | 0.67 |
ENSDART00000018071
|
eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr1_+_55703120 | 0.67 |
ENSDART00000141089
|
adgre6
|
adhesion G protein-coupled receptor E6 |
| chr2_+_3452590 | 0.67 |
ENSDART00000153935
|
cyp8b2
|
cytochrome P450, family 8, subfamily B, polypeptide 2 |
| chr1_+_277731 | 0.65 |
ENSDART00000133431
|
cenpe
|
centromere protein E |
| chr8_-_1219815 | 0.65 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr5_+_25762271 | 0.65 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr20_-_26929685 | 0.62 |
ENSDART00000132556
|
ftr79
|
finTRIM family, member 79 |
| chr9_-_105135 | 0.62 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr3_-_7990516 | 0.62 |
ENSDART00000167877
|
TRIM35 (1 of many)
|
si:ch211-175l6.2 |
| chr3_-_180860 | 0.62 |
ENSDART00000059956
ENSDART00000192506 |
kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr22_+_5971404 | 0.61 |
ENSDART00000122153
|
BX322587.1
|
|
| chr5_-_65000312 | 0.61 |
ENSDART00000192893
|
ANXA1 (1 of many)
|
zgc:110283 |
| chr7_+_26029672 | 0.61 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr1_-_513762 | 0.61 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr3_-_34098731 | 0.61 |
ENSDART00000150999
|
ighv5-3
|
immunoglobulin heavy variable 5-3 |
| chr17_-_2573021 | 0.61 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr16_+_9495583 | 0.60 |
ENSDART00000150750
ENSDART00000150457 |
pimr208
|
Pim proto-oncogene, serine/threonine kinase, related 208 |
| chr25_+_25202213 | 0.60 |
ENSDART00000150744
|
ccl39a.10
|
chemokine (C-C motif) ligand 39, duplicate 10 |
| chr19_-_3783141 | 0.60 |
ENSDART00000162260
|
btr19
|
bloodthirsty-related gene family, member 19 |
| chr11_+_42587900 | 0.59 |
ENSDART00000167529
|
asb14a
|
ankyrin repeat and SOCS box containing 14a |
| chr6_-_35439406 | 0.59 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr22_+_36704764 | 0.59 |
ENSDART00000146662
|
trim35-33
|
tripartite motif containing 35-33 |
| chr2_-_44255537 | 0.58 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr8_-_1051438 | 0.58 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr7_-_8470860 | 0.58 |
ENSDART00000172793
|
loc564481
|
hypothetical protein LOC564481 |
| chr15_+_38299563 | 0.58 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr17_-_2578026 | 0.58 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr18_+_5547185 | 0.57 |
ENSDART00000193977
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr7_-_6346859 | 0.57 |
ENSDART00000172913
|
si:ch73-368j24.11
|
si:ch73-368j24.11 |
| chr22_-_10541712 | 0.56 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr1_+_55002583 | 0.56 |
ENSDART00000037250
|
si:ch211-196h16.12
|
si:ch211-196h16.12 |
| chr9_+_44994214 | 0.55 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr4_-_58964138 | 0.55 |
ENSDART00000150259
|
si:ch211-64i20.3
|
si:ch211-64i20.3 |
| chr6_-_13206255 | 0.55 |
ENSDART00000065373
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr3_+_2669813 | 0.55 |
ENSDART00000014205
|
CR388047.1
|
|
| chr17_+_53439606 | 0.54 |
ENSDART00000154946
|
si:zfos-1714f5.3
|
si:zfos-1714f5.3 |
| chr3_-_4760384 | 0.54 |
ENSDART00000108810
|
CABZ01046997.1
|
|
| chr1_+_55452892 | 0.54 |
ENSDART00000122508
|
CR788255.1
|
|
| chr18_-_11729 | 0.53 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr19_-_27588842 | 0.53 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr15_+_47746176 | 0.53 |
ENSDART00000154481
ENSDART00000160914 |
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr1_-_9644630 | 0.53 |
ENSDART00000123725
ENSDART00000161164 |
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr17_-_2590222 | 0.53 |
ENSDART00000185711
|
CR759892.1
|
|
| chr9_+_33217243 | 0.52 |
ENSDART00000053061
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr10_+_40629616 | 0.52 |
ENSDART00000147476
|
CR396590.9
|
|
| chr24_-_30275735 | 0.52 |
ENSDART00000168723
|
snx7
|
sorting nexin 7 |
| chr21_-_31210749 | 0.51 |
ENSDART00000185356
|
zgc:152891
|
zgc:152891 |
| chr3_+_1179601 | 0.51 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr4_-_77636185 | 0.51 |
ENSDART00000152915
ENSDART00000131964 |
si:dkey-61p9.9
|
si:dkey-61p9.9 |
| chr1_+_292545 | 0.50 |
ENSDART00000148261
|
cenpe
|
centromere protein E |
| chr2_-_51757328 | 0.50 |
ENSDART00000189286
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr21_+_53504 | 0.50 |
ENSDART00000170452
|
dmgdh
|
dimethylglycine dehydrogenase |
| chr25_+_37289088 | 0.50 |
ENSDART00000073438
|
si:dkey-234i14.9
|
si:dkey-234i14.9 |
| chr4_+_77709678 | 0.49 |
ENSDART00000036856
|
AL935186.1
|
|
| chr5_-_7829657 | 0.49 |
ENSDART00000158374
|
pdlim5a
|
PDZ and LIM domain 5a |
| chr1_-_53880639 | 0.49 |
ENSDART00000010543
|
ltv1
|
LTV1 ribosome biogenesis factor |
| chr4_-_32456788 | 0.49 |
ENSDART00000151862
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr25_+_37290206 | 0.49 |
ENSDART00000086474
|
si:dkey-234i14.12
|
si:dkey-234i14.12 |
| chr13_-_42560662 | 0.48 |
ENSDART00000124898
|
CR792417.1
|
|
| chr7_+_8358547 | 0.48 |
ENSDART00000173016
|
jac7
|
jacalin 7 |
| chr4_+_50401133 | 0.48 |
ENSDART00000150644
|
si:dkey-156k2.7
|
si:dkey-156k2.7 |
| chr22_+_18469004 | 0.48 |
ENSDART00000061430
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr25_-_37501371 | 0.47 |
ENSDART00000160498
|
CABZ01088346.1
|
|
| chr21_-_30293224 | 0.47 |
ENSDART00000101051
|
slbp2
|
stem-loop binding protein 2 |
| chr3_-_29977495 | 0.47 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr22_+_9262800 | 0.47 |
ENSDART00000143041
|
si:ch211-250k18.6
|
si:ch211-250k18.6 |
| chr16_+_42772678 | 0.46 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr25_-_36369057 | 0.46 |
ENSDART00000064400
|
si:ch211-113a14.24
|
si:ch211-113a14.24 |
| chr1_+_50293938 | 0.46 |
ENSDART00000084184
|
aimp1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr21_-_44081540 | 0.46 |
ENSDART00000130833
|
FO704810.1
|
|
| chr23_+_29908348 | 0.45 |
ENSDART00000186622
ENSDART00000150174 |
si:ch73-236e11.2
|
si:ch73-236e11.2 |
| chr3_+_4726621 | 0.45 |
ENSDART00000184186
|
CABZ01046996.1
|
|
| chr12_-_44151296 | 0.44 |
ENSDART00000168734
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr1_+_57235896 | 0.44 |
ENSDART00000152621
|
si:dkey-27j5.7
|
si:dkey-27j5.7 |
| chr9_+_32178050 | 0.44 |
ENSDART00000169526
|
coq10b
|
coenzyme Q10B |
| chr5_-_26181863 | 0.44 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr2_-_59345920 | 0.44 |
ENSDART00000134662
|
ftr37
|
finTRIM family, member 37 |
| chr8_+_39570615 | 0.44 |
ENSDART00000142557
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr21_+_2203316 | 0.43 |
ENSDART00000185659
|
BX510922.4
|
|
| chr14_+_17382803 | 0.43 |
ENSDART00000040383
|
si:ch211-255i20.3
|
si:ch211-255i20.3 |
| chr20_-_54377933 | 0.43 |
ENSDART00000182664
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr11_+_37909654 | 0.43 |
ENSDART00000172211
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr5_+_37744625 | 0.43 |
ENSDART00000014031
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr23_-_45319592 | 0.42 |
ENSDART00000189861
|
ccdc171
|
coiled-coil domain containing 171 |
| chr7_-_6415991 | 0.42 |
ENSDART00000173349
|
CU457819.3
|
Histone H3.2 |
| chr1_+_56823437 | 0.42 |
ENSDART00000129527
|
si:ch211-1f22.5
|
si:ch211-1f22.5 |
| chr13_-_50565338 | 0.42 |
ENSDART00000062684
|
blnk
|
B cell linker |
| chr17_+_132555 | 0.41 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr5_-_30984010 | 0.41 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr14_-_25928899 | 0.41 |
ENSDART00000143518
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr2_-_32624577 | 0.41 |
ENSDART00000112797
|
si:dkeyp-73d8.6
|
si:dkeyp-73d8.6 |
| chr16_-_19568795 | 0.41 |
ENSDART00000185141
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr3_-_61203203 | 0.41 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr7_+_5964296 | 0.41 |
ENSDART00000173380
|
si:dkey-23a13.17
|
si:dkey-23a13.17 |
| chr16_-_48914757 | 0.41 |
ENSDART00000166740
|
FQ976914.1
|
|
| chr20_+_25225112 | 0.40 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr8_+_28469054 | 0.40 |
ENSDART00000062716
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr23_-_18057270 | 0.40 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr10_+_4987766 | 0.40 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr17_-_7457698 | 0.40 |
ENSDART00000144492
ENSDART00000172035 |
si:ch211-278p9.1
|
si:ch211-278p9.1 |
| chr4_+_2267641 | 0.40 |
ENSDART00000165503
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr24_+_5789582 | 0.39 |
ENSDART00000141504
|
BX470065.1
|
|
| chr7_+_33132074 | 0.39 |
ENSDART00000073554
|
zgc:153219
|
zgc:153219 |
| chr6_-_51573975 | 0.39 |
ENSDART00000073865
|
rbl1
|
retinoblastoma-like 1 (p107) |
| chr7_+_25053331 | 0.39 |
ENSDART00000173998
|
si:dkey-23i12.7
|
si:dkey-23i12.7 |
| chr7_+_56577906 | 0.39 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr8_+_40275830 | 0.39 |
ENSDART00000164414
|
orai1a
|
ORAI calcium release-activated calcium modulator 1a |
| chr15_-_551177 | 0.39 |
ENSDART00000066774
ENSDART00000154617 |
tagln
|
transgelin |
| chr18_+_5454341 | 0.39 |
ENSDART00000192649
|
dtwd1
|
DTW domain containing 1 |
| chr7_+_35229645 | 0.39 |
ENSDART00000144327
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr12_-_44148073 | 0.38 |
ENSDART00000166273
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr1_-_56717813 | 0.38 |
ENSDART00000159196
|
FO681314.1
|
|
| chr7_-_6470431 | 0.38 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr19_-_10432134 | 0.38 |
ENSDART00000081440
|
il11b
|
interleukin 11b |
| chr4_+_37952218 | 0.38 |
ENSDART00000186865
|
si:dkeyp-82b4.2
|
si:dkeyp-82b4.2 |
| chr17_+_53156530 | 0.38 |
ENSDART00000126277
ENSDART00000156774 |
dph6
|
diphthamine biosynthesis 6 |
| chr5_-_8907819 | 0.38 |
ENSDART00000188523
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr1_-_55118745 | 0.38 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr7_+_24523017 | 0.38 |
ENSDART00000077047
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr17_+_20607487 | 0.37 |
ENSDART00000154123
|
si:ch73-288o11.5
|
si:ch73-288o11.5 |
| chr4_-_43911462 | 0.37 |
ENSDART00000189056
ENSDART00000162441 ENSDART00000171367 |
znf1104
|
zinc finger protein 1104 |
| chr2_-_59265521 | 0.37 |
ENSDART00000146341
ENSDART00000097799 |
ftr33
|
finTRIM family, member 33 |
| chr7_-_7984015 | 0.37 |
ENSDART00000163203
|
si:cabz01030277.1
|
si:cabz01030277.1 |
| chr22_+_38310957 | 0.37 |
ENSDART00000040550
|
traf5
|
Tnf receptor-associated factor 5 |
| chr15_-_4485828 | 0.36 |
ENSDART00000062868
|
tfdp2
|
transcription factor Dp-2 |
| chr4_-_65747476 | 0.36 |
ENSDART00000142421
|
si:dkey-28k24.2
|
si:dkey-28k24.2 |
| chr5_-_15692030 | 0.36 |
ENSDART00000099545
|
CR384085.1
|
|
| chr2_-_59247811 | 0.36 |
ENSDART00000141384
|
ftr32
|
finTRIM family, member 32 |
| chr4_+_8638622 | 0.36 |
ENSDART00000186829
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr17_+_1627379 | 0.36 |
ENSDART00000184050
|
LO018432.1
|
|
| chr2_-_36497113 | 0.36 |
ENSDART00000109291
|
BX901889.4
|
|
| chr19_-_24555935 | 0.36 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr4_+_75577480 | 0.35 |
ENSDART00000188196
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr6_+_27315173 | 0.35 |
ENSDART00000108831
|
espnla
|
espin like a |
| chr3_-_45848043 | 0.35 |
ENSDART00000055132
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of tal1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.3 | 1.0 | GO:0042148 | strand invasion(GO:0042148) |
| 0.3 | 0.8 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 0.6 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.2 | 0.9 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.2 | 0.7 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.2 | 0.8 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.2 | 2.8 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.7 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.6 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 2.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.0 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.1 | 0.4 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.4 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 0.3 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.1 | 1.0 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.3 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 0.8 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 1.1 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.3 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 1.8 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 2.0 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.1 | 0.4 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 1.1 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.4 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.5 | GO:0040016 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) embryonic cleavage(GO:0040016) |
| 0.1 | 0.3 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 0.4 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 0.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.4 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.3 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.3 | GO:1990592 | IRE1-mediated unfolded protein response(GO:0036498) negative regulation of response to endoplasmic reticulum stress(GO:1903573) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.6 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 2.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.5 | GO:0003211 | cardiac ventricle formation(GO:0003211) |
| 0.1 | 0.3 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.1 | 0.3 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.2 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.0 | 0.2 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.0 | 1.4 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 1.7 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.2 | GO:1904396 | regulation of neuromuscular junction development(GO:1904396) |
| 0.0 | 0.1 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.3 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 1.2 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.4 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.4 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.6 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.7 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 1.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.4 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.0 | 0.2 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.4 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.2 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 1.7 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.0 | GO:0019264 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.4 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.3 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.2 | 1.0 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.3 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.1 | 0.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.6 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 2.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.6 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.7 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.6 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 1.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 2.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.0 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.2 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.3 | GO:0000784 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.4 | GO:0043186 | P granule(GO:0043186) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 0.7 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.2 | 0.7 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.2 | 0.8 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.2 | 0.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 0.5 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.2 | 1.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 0.6 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.6 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.4 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.5 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 0.8 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 1.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 2.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.3 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 0.2 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.1 | 0.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.4 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 1.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.0 | GO:0022884 | macromolecule transmembrane transporter activity(GO:0022884) |
| 0.1 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.6 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 2.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.6 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 1.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 1.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.4 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.2 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.1 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.0 | 0.4 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.2 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.2 | GO:0030250 | calcium sensitive guanylate cyclase activator activity(GO:0008048) cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.0 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.2 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |