Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
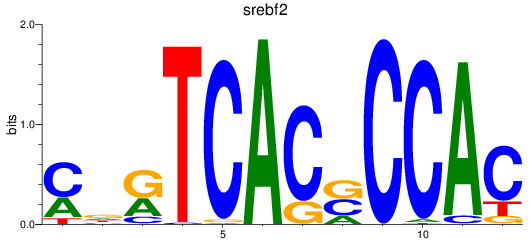
Results for srebf2
Z-value: 1.15
Transcription factors associated with srebf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
srebf2
|
ENSDARG00000063438 | sterol regulatory element binding transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| srebf2 | dr11_v1_chr3_+_1107102_1107102 | -0.06 | 9.3e-01 | Click! |
Activity profile of srebf2 motif
Sorted Z-values of srebf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_7539297 | 0.75 |
ENSDART00000081550
ENSDART00000081553 |
desma
|
desmin a |
| chr4_-_20511595 | 0.74 |
ENSDART00000185806
|
rassf8b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8b |
| chr19_-_25113660 | 0.74 |
ENSDART00000035538
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr21_+_28445052 | 0.67 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr10_-_31782616 | 0.64 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr18_-_21271373 | 0.62 |
ENSDART00000060001
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr14_-_21618005 | 0.62 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr16_+_41873708 | 0.61 |
ENSDART00000084631
ENSDART00000084639 ENSDART00000058611 |
scn1ba
|
sodium channel, voltage-gated, type I, beta a |
| chr13_-_33398735 | 0.58 |
ENSDART00000182601
ENSDART00000103628 |
btbd6a
|
BTB (POZ) domain containing 6a |
| chr7_+_44445595 | 0.58 |
ENSDART00000108766
|
cdh5
|
cadherin 5 |
| chr14_-_29826659 | 0.57 |
ENSDART00000138413
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr22_-_11614973 | 0.55 |
ENSDART00000063135
|
pcyt2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr23_-_20258490 | 0.55 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr21_-_19006631 | 0.52 |
ENSDART00000080269
ENSDART00000191682 |
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr2_+_24203229 | 0.51 |
ENSDART00000138088
|
map4l
|
microtubule associated protein 4 like |
| chr9_-_4075620 | 0.50 |
ENSDART00000046903
|
mylka
|
myosin, light chain kinase a |
| chr16_-_29164379 | 0.49 |
ENSDART00000132589
|
mef2d
|
myocyte enhancer factor 2d |
| chr20_-_27311675 | 0.47 |
ENSDART00000026088
ENSDART00000148361 |
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr21_-_43550120 | 0.46 |
ENSDART00000151627
|
si:ch73-362m14.2
|
si:ch73-362m14.2 |
| chr4_+_19535946 | 0.45 |
ENSDART00000192342
ENSDART00000183740 ENSDART00000180812 ENSDART00000180017 |
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr8_-_26961779 | 0.44 |
ENSDART00000099214
|
slc16a1b
|
solute carrier family 16 (monocarboxylate transporter), member 1b |
| chr3_-_61099004 | 0.41 |
ENSDART00000112043
|
cacng4b
|
calcium channel, voltage-dependent, gamma subunit 4b |
| chr1_+_41690402 | 0.41 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr12_-_23009312 | 0.41 |
ENSDART00000111801
|
mkxa
|
mohawk homeobox a |
| chr9_-_43082945 | 0.41 |
ENSDART00000142257
|
ccdc141
|
coiled-coil domain containing 141 |
| chr17_-_48915427 | 0.40 |
ENSDART00000054781
|
lgals8b
|
galectin 8b |
| chr24_+_9590188 | 0.39 |
ENSDART00000137092
|
si:dkey-96n2.1
|
si:dkey-96n2.1 |
| chr22_-_13466246 | 0.35 |
ENSDART00000134035
|
cntnap5b
|
contactin associated protein-like 5b |
| chr17_+_15041647 | 0.34 |
ENSDART00000108999
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr3_+_26288981 | 0.32 |
ENSDART00000163500
|
rhot1a
|
ras homolog family member T1a |
| chr8_+_52491436 | 0.32 |
ENSDART00000164298
|
si:ch1073-392o20.1
|
si:ch1073-392o20.1 |
| chr3_-_48612078 | 0.30 |
ENSDART00000169923
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr19_-_10196370 | 0.29 |
ENSDART00000091707
|
dbpa
|
D site albumin promoter binding protein a |
| chr14_-_33894915 | 0.28 |
ENSDART00000143290
|
urp1
|
urotensin-related peptide 1 |
| chr9_+_54417141 | 0.27 |
ENSDART00000056810
|
drd1b
|
dopamine receptor D1b |
| chr7_-_30082931 | 0.25 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr10_-_35220285 | 0.25 |
ENSDART00000180439
|
ypel2a
|
yippee-like 2a |
| chr5_+_45976130 | 0.24 |
ENSDART00000175670
|
sv2c
|
synaptic vesicle glycoprotein 2C |
| chr17_-_7436766 | 0.23 |
ENSDART00000162002
|
grm1b
|
glutamate receptor, metabotropic 1b |
| chr5_-_36328688 | 0.20 |
ENSDART00000011399
|
efnb1
|
ephrin-B1 |
| chr5_-_37871526 | 0.19 |
ENSDART00000136450
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr8_-_13678415 | 0.18 |
ENSDART00000134153
ENSDART00000143331 |
si:dkey-258f14.3
|
si:dkey-258f14.3 |
| chr10_-_31563049 | 0.18 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr14_-_46117345 | 0.18 |
ENSDART00000172166
|
NDUFC1
|
si:ch211-235e9.6 |
| chr13_-_992458 | 0.18 |
ENSDART00000114655
|
BFSP1
|
beaded filament structural protein 1 |
| chr18_-_39188664 | 0.18 |
ENSDART00000162983
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr1_-_30689004 | 0.17 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr12_+_33395748 | 0.16 |
ENSDART00000129458
|
fasn
|
fatty acid synthase |
| chr5_-_12524996 | 0.16 |
ENSDART00000142258
|
si:ch73-263f13.1
|
si:ch73-263f13.1 |
| chr3_-_45308394 | 0.15 |
ENSDART00000155324
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr13_+_4409294 | 0.15 |
ENSDART00000146437
|
si:ch211-130h14.4
|
si:ch211-130h14.4 |
| chr6_-_19351495 | 0.15 |
ENSDART00000164287
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr16_+_41874001 | 0.15 |
ENSDART00000161278
|
scn1ba
|
sodium channel, voltage-gated, type I, beta a |
| chr3_-_34296361 | 0.15 |
ENSDART00000181485
ENSDART00000113726 |
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr18_+_8231138 | 0.14 |
ENSDART00000140193
|
arsa
|
arylsulfatase A |
| chr19_+_20785724 | 0.13 |
ENSDART00000038942
|
adnp2b
|
ADNP homeobox 2b |
| chr6_-_15757867 | 0.13 |
ENSDART00000063665
|
ackr3b
|
atypical chemokine receptor 3b |
| chr11_-_3334248 | 0.13 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr7_-_41013575 | 0.12 |
ENSDART00000150139
|
insig1
|
insulin induced gene 1 |
| chr1_+_9557212 | 0.12 |
ENSDART00000111131
|
elfn1b
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1b |
| chr25_+_3008263 | 0.12 |
ENSDART00000160484
|
neo1b
|
neogenin 1b |
| chr6_-_15101477 | 0.11 |
ENSDART00000187713
ENSDART00000124132 |
fhl2b
|
four and a half LIM domains 2b |
| chr18_+_39649825 | 0.11 |
ENSDART00000087960
|
gldn
|
gliomedin |
| chr20_+_39223235 | 0.10 |
ENSDART00000132132
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr13_+_30572172 | 0.10 |
ENSDART00000010052
ENSDART00000144417 |
ppifa
|
peptidylprolyl isomerase Fa |
| chr9_-_30363770 | 0.09 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr4_+_646154 | 0.09 |
ENSDART00000171138
|
xpot
|
exportin, tRNA (nuclear export receptor for tRNAs) |
| chr3_-_6417328 | 0.09 |
ENSDART00000160979
|
jpt1b
|
Jupiter microtubule associated homolog 1b |
| chr23_-_29878643 | 0.08 |
ENSDART00000058407
|
slc25a33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr8_+_4745340 | 0.08 |
ENSDART00000182894
|
ufd1l
|
ubiquitin recognition factor in ER associated degradation 1 |
| chr10_-_5053589 | 0.08 |
ENSDART00000193579
|
tmem150c
|
transmembrane protein 150C |
| chr4_+_41311345 | 0.07 |
ENSDART00000151905
|
si:dkey-16p19.8
|
si:dkey-16p19.8 |
| chr23_-_27694960 | 0.07 |
ENSDART00000135129
|
IKZF4
|
si:dkey-166n8.9 |
| chr22_+_17286841 | 0.07 |
ENSDART00000133176
|
FAM163A
|
si:dkey-171o17.8 |
| chr5_+_37032038 | 0.06 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr3_-_51109286 | 0.06 |
ENSDART00000172010
|
si:ch211-148f13.1
|
si:ch211-148f13.1 |
| chr5_-_14344647 | 0.06 |
ENSDART00000188456
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr2_+_34138376 | 0.05 |
ENSDART00000137170
|
dars2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr20_-_7176809 | 0.05 |
ENSDART00000012247
ENSDART00000125019 |
dhcr24
|
24-dehydrocholesterol reductase |
| chr14_-_30390145 | 0.05 |
ENSDART00000045423
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr7_-_5640822 | 0.05 |
ENSDART00000143767
|
BX000524.1
|
|
| chr8_+_22916536 | 0.05 |
ENSDART00000136403
|
cacna1fb
|
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr18_+_14342326 | 0.04 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr2_-_48539673 | 0.04 |
ENSDART00000168202
|
CR391991.4
|
|
| chr22_-_13165186 | 0.04 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr7_+_25920792 | 0.03 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr5_-_15321451 | 0.03 |
ENSDART00000139203
|
txnrd2.1
|
thioredoxin reductase 2, tandem duplicate 1 |
| chr24_-_3477103 | 0.02 |
ENSDART00000143723
|
idi1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr13_+_21826096 | 0.02 |
ENSDART00000056432
|
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr12_+_21684239 | 0.01 |
ENSDART00000043546
|
mrpl27
|
mitochondrial ribosomal protein L27 |
| chr24_-_28229618 | 0.01 |
ENSDART00000145290
|
bcl2a
|
BCL2, apoptosis regulator a |
Network of associatons between targets according to the STRING database.
First level regulatory network of srebf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.2 | 0.7 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.1 | 0.8 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.1 | 0.6 | GO:0001955 | blood vessel maturation(GO:0001955) regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.7 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0036314 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.3 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.5 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.3 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.1 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.0 | 0.5 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.1 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.2 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.0 | 0.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.4 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.4 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.4 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.1 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.6 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.5 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.3 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.0 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |