Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
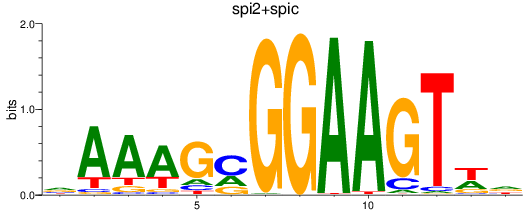
Results for spi2+spic
Z-value: 4.34
Transcription factors associated with spi2+spic
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
spic
|
ENSDARG00000012435 | Spi-C transcription factor (Spi-1/PU.1 related) |
|
spi2
|
ENSDARG00000087438 | Spi-2 proto-oncogene |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| spic | dr11_v1_chr4_-_17812131_17812131 | 0.78 | 1.2e-01 | Click! |
Activity profile of spi2+spic motif
Sorted Z-values of spi2+spic motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_18626515 | 3.18 |
ENSDART00000160624
|
rps18
|
ribosomal protein S18 |
| chr16_+_20934353 | 2.88 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr5_-_25582721 | 2.83 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr4_-_22310956 | 2.79 |
ENSDART00000162585
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr5_-_25583125 | 2.73 |
ENSDART00000031665
ENSDART00000145353 |
anxa1a
|
annexin A1a |
| chr10_-_15128771 | 2.70 |
ENSDART00000101261
|
spp1
|
secreted phosphoprotein 1 |
| chr22_+_5478353 | 2.68 |
ENSDART00000160596
|
tppp
|
tubulin polymerization promoting protein |
| chr9_+_21165017 | 2.52 |
ENSDART00000145933
ENSDART00000142985 |
si:rp71-68n21.9
|
si:rp71-68n21.9 |
| chr3_+_29510818 | 2.45 |
ENSDART00000055407
ENSDART00000193743 ENSDART00000123619 |
rac2
|
Rac family small GTPase 2 |
| chr23_+_39089574 | 2.43 |
ENSDART00000164711
|
nfatc2a
|
nuclear factor of activated T cells 2a |
| chr14_+_36886950 | 2.37 |
ENSDART00000183719
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr2_+_24762567 | 2.31 |
ENSDART00000078866
|
ifi30
|
interferon, gamma-inducible protein 30 |
| chr21_+_27448856 | 2.26 |
ENSDART00000100784
|
cfbl
|
complement factor b-like |
| chr21_-_22720381 | 2.23 |
ENSDART00000130789
|
c1qc
|
complement component 1, q subcomponent, C chain |
| chr1_+_52735484 | 2.22 |
ENSDART00000182076
|
CABZ01021532.1
|
|
| chr16_+_31542645 | 2.19 |
ENSDART00000163724
|
SLA (1 of many)
|
Src like adaptor |
| chr2_-_40199780 | 2.17 |
ENSDART00000113901
|
ccl34a.4
|
chemokine (C-C motif) ligand 34a, duplicate 4 |
| chr13_+_23623346 | 2.16 |
ENSDART00000057619
|
il22ra2
|
interleukin 22 receptor, alpha 2 |
| chr3_+_36424055 | 2.13 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr7_-_56606752 | 2.12 |
ENSDART00000138714
|
sult5a1
|
sulfotransferase family 5A, member 1 |
| chr5_+_37087583 | 2.11 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr3_-_57744323 | 2.10 |
ENSDART00000101829
|
lgals3bpb
|
lectin, galactoside-binding, soluble, 3 binding protein b |
| chr1_+_57787642 | 2.09 |
ENSDART00000127091
|
si:dkey-1c7.2
|
si:dkey-1c7.2 |
| chr20_-_14054083 | 2.09 |
ENSDART00000009549
|
rhag
|
Rh associated glycoprotein |
| chr25_+_13205878 | 2.08 |
ENSDART00000162319
ENSDART00000162283 |
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr10_+_42358426 | 2.03 |
ENSDART00000025691
|
dbnla
|
drebrin-like a |
| chr10_+_41159241 | 2.00 |
ENSDART00000141657
|
anxa4
|
annexin A4 |
| chr4_-_22311610 | 1.95 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr8_-_54077740 | 1.95 |
ENSDART00000027000
|
rho
|
rhodopsin |
| chr25_+_13191615 | 1.94 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr8_-_2153147 | 1.90 |
ENSDART00000124093
|
si:dkeyp-117b11.1
|
si:dkeyp-117b11.1 |
| chr15_+_46356879 | 1.90 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr3_-_32965848 | 1.86 |
ENSDART00000050930
|
casp6
|
caspase 6, apoptosis-related cysteine peptidase |
| chr13_+_31716820 | 1.81 |
ENSDART00000034745
|
prkcha
|
protein kinase C, eta, a |
| chr22_+_5687615 | 1.78 |
ENSDART00000133241
ENSDART00000019854 ENSDART00000138102 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr20_+_26880668 | 1.78 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr24_-_27436319 | 1.77 |
ENSDART00000171489
|
si:dkey-25o1.7
|
si:dkey-25o1.7 |
| chr22_-_31020690 | 1.75 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr4_-_11603149 | 1.74 |
ENSDART00000150468
|
net1
|
neuroepithelial cell transforming 1 |
| chr20_+_36812368 | 1.73 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr13_+_228045 | 1.70 |
ENSDART00000161091
|
zgc:64201
|
zgc:64201 |
| chr20_+_16881883 | 1.69 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr25_+_13191391 | 1.69 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr5_+_22510639 | 1.66 |
ENSDART00000080919
|
rpl36a
|
ribosomal protein L36A |
| chr17_+_33415319 | 1.63 |
ENSDART00000140805
ENSDART00000025501 ENSDART00000146447 |
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr12_-_30558694 | 1.63 |
ENSDART00000153417
|
si:ch211-28p3.3
|
si:ch211-28p3.3 |
| chr17_-_43022468 | 1.62 |
ENSDART00000002945
|
NPC2 (1 of many)
|
zgc:193725 |
| chr23_-_40766518 | 1.60 |
ENSDART00000127420
|
si:dkeyp-27c8.2
|
si:dkeyp-27c8.2 |
| chr9_-_54304684 | 1.59 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr19_+_4038589 | 1.57 |
ENSDART00000169271
|
btr23
|
bloodthirsty-related gene family, member 23 |
| chr9_+_21165484 | 1.57 |
ENSDART00000177286
|
si:rp71-68n21.9
|
si:rp71-68n21.9 |
| chr17_-_48705993 | 1.56 |
ENSDART00000030934
|
kcnk5a
|
potassium channel, subfamily K, member 5a |
| chr5_+_24086227 | 1.56 |
ENSDART00000051549
ENSDART00000177458 ENSDART00000135934 |
tp53
|
tumor protein p53 |
| chr15_-_2652640 | 1.55 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr22_+_36719821 | 1.52 |
ENSDART00000149093
|
TRIM35 (1 of many)
|
si:ch1073-324l1.1 |
| chr18_+_31073265 | 1.51 |
ENSDART00000023539
|
cyba
|
cytochrome b-245, alpha polypeptide |
| chr10_-_7785930 | 1.49 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr24_-_26479841 | 1.49 |
ENSDART00000079984
|
rpl22l1
|
ribosomal protein L22-like 1 |
| chr24_-_36680261 | 1.49 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr3_-_58831683 | 1.48 |
ENSDART00000110292
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr4_-_18954001 | 1.47 |
ENSDART00000144814
|
si:dkey-31f5.8
|
si:dkey-31f5.8 |
| chr20_-_34127415 | 1.43 |
ENSDART00000010028
|
ptgs2b
|
prostaglandin-endoperoxide synthase 2b |
| chr1_+_57774454 | 1.43 |
ENSDART00000152507
|
si:dkey-1c7.2
|
si:dkey-1c7.2 |
| chr9_-_28687484 | 1.42 |
ENSDART00000136537
|
si:ch73-7i4.3
|
si:ch73-7i4.3 |
| chr2_+_1945211 | 1.41 |
ENSDART00000109668
|
bcl10
|
B cell CLL/lymphoma 10 |
| chr14_-_706345 | 1.40 |
ENSDART00000166632
|
si:zfos-741a10.3
|
si:zfos-741a10.3 |
| chr10_+_35422802 | 1.38 |
ENSDART00000147303
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr24_+_264839 | 1.36 |
ENSDART00000066872
|
ccr12b.2
|
chemokine (C-C motif) receptor 12b, tandem duplicate 2 |
| chr22_+_19486711 | 1.35 |
ENSDART00000144389
ENSDART00000139256 ENSDART00000160855 |
si:dkey-78l4.10
|
si:dkey-78l4.10 |
| chr7_-_21925863 | 1.35 |
ENSDART00000191213
|
si:dkey-85k7.10
|
si:dkey-85k7.10 |
| chr14_+_10505464 | 1.33 |
ENSDART00000091224
|
p2ry10
|
P2Y receptor family member 10 |
| chr3_-_16719244 | 1.33 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr14_-_5690363 | 1.32 |
ENSDART00000144539
|
tlx2
|
T cell leukemia homeobox 2 |
| chr21_-_7035599 | 1.30 |
ENSDART00000139777
|
si:ch211-93g21.1
|
si:ch211-93g21.1 |
| chr19_-_18626952 | 1.30 |
ENSDART00000168004
ENSDART00000162034 ENSDART00000165486 ENSDART00000167971 |
rps18
|
ribosomal protein S18 |
| chr19_+_7045033 | 1.29 |
ENSDART00000146579
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr3_-_32603191 | 1.29 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr7_+_19600262 | 1.28 |
ENSDART00000007310
|
zgc:171731
|
zgc:171731 |
| chr7_+_26545911 | 1.27 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr3_-_34100700 | 1.27 |
ENSDART00000151628
|
ighv6-1
|
immunoglobulin heavy variable 6-1 |
| chr21_+_76739 | 1.26 |
ENSDART00000174654
|
ARSB
|
arylsulfatase B |
| chr17_-_6451801 | 1.26 |
ENSDART00000064700
|
fuca2
|
alpha-L-fucosidase 2 |
| chr16_-_22303130 | 1.25 |
ENSDART00000142181
|
si:dkey-92i15.4
|
si:dkey-92i15.4 |
| chr3_-_30941362 | 1.24 |
ENSDART00000076830
|
coro1a
|
coronin, actin binding protein, 1A |
| chr8_-_29713595 | 1.24 |
ENSDART00000131988
ENSDART00000077637 |
mpeg1.1
|
macrophage expressed 1, tandem duplicate 1 |
| chr1_-_25177086 | 1.23 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr19_+_348729 | 1.22 |
ENSDART00000114284
|
mcl1a
|
MCL1, BCL2 family apoptosis regulator a |
| chr13_+_159504 | 1.22 |
ENSDART00000057248
|
ewsr1b
|
EWS RNA-binding protein 1b |
| chr3_+_18579806 | 1.22 |
ENSDART00000180967
ENSDART00000089765 |
arhgap17b
|
Rho GTPase activating protein 17b |
| chr5_+_23913585 | 1.22 |
ENSDART00000015401
|
ercc6l
|
excision repair cross-complementation group 6-like |
| chr4_+_76637548 | 1.21 |
ENSDART00000133799
|
ms4a17a.9
|
membrane-spanning 4-domains, subfamily A, member 17A.9 |
| chr9_-_30555725 | 1.21 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr17_+_33415542 | 1.20 |
ENSDART00000183169
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr14_-_11529311 | 1.17 |
ENSDART00000127208
|
si:ch211-153b23.7
|
si:ch211-153b23.7 |
| chr7_-_52417777 | 1.17 |
ENSDART00000110265
|
myzap
|
myocardial zonula adherens protein |
| chr20_+_305035 | 1.17 |
ENSDART00000104807
|
si:dkey-119m7.4
|
si:dkey-119m7.4 |
| chr11_+_21096339 | 1.16 |
ENSDART00000124574
|
il19l
|
interleukin 19 like |
| chr14_-_40797117 | 1.15 |
ENSDART00000122369
|
elf1
|
E74-like ETS transcription factor 1 |
| chr16_+_38277339 | 1.15 |
ENSDART00000085143
|
bnipl
|
BCL2 interacting protein like |
| chr11_+_29975830 | 1.15 |
ENSDART00000148929
|
si:ch73-226l13.2
|
si:ch73-226l13.2 |
| chr18_+_36631923 | 1.15 |
ENSDART00000098980
|
znf296
|
zinc finger protein 296 |
| chr1_-_29658721 | 1.15 |
ENSDART00000132063
|
si:dkey-1h24.6
|
si:dkey-1h24.6 |
| chr15_-_29586747 | 1.14 |
ENSDART00000076749
|
samsn1a
|
SAM domain, SH3 domain and nuclear localisation signals 1a |
| chr16_+_29514473 | 1.14 |
ENSDART00000034102
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr4_+_76745015 | 1.14 |
ENSDART00000155883
|
ms4a17a.10
|
membrane-spanning 4-domains, subfamily A, member 17A.10 |
| chr12_+_27536270 | 1.13 |
ENSDART00000133719
|
etv4
|
ets variant 4 |
| chr25_-_34280080 | 1.13 |
ENSDART00000085251
|
gcnt3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr21_+_30937690 | 1.12 |
ENSDART00000022562
|
rhogb
|
ras homolog family member Gb |
| chr22_-_4769140 | 1.11 |
ENSDART00000165235
|
calr3a
|
calreticulin 3a |
| chr10_-_33557299 | 1.11 |
ENSDART00000099732
|
il2rga
|
interleukin 2 receptor, gamma a |
| chr1_-_9228007 | 1.10 |
ENSDART00000147277
ENSDART00000135219 |
gng13a
|
guanine nucleotide binding protein (G protein), gamma 13a |
| chr5_-_51166025 | 1.10 |
ENSDART00000097471
|
card9
|
caspase recruitment domain family, member 9 |
| chr21_-_26114886 | 1.10 |
ENSDART00000139320
|
nipal4
|
NIPA-like domain containing 4 |
| chr11_+_29965822 | 1.09 |
ENSDART00000127049
|
il1fma
|
interleukin-1 family member A |
| chr7_+_32901658 | 1.08 |
ENSDART00000115420
|
ano9b
|
anoctamin 9b |
| chr19_-_40175800 | 1.08 |
ENSDART00000102904
|
illr4
|
immune-related, lectin-like receptor 4 |
| chr7_+_49664174 | 1.08 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr16_-_31756859 | 1.08 |
ENSDART00000149170
ENSDART00000126617 ENSDART00000182722 |
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr7_-_50764714 | 1.07 |
ENSDART00000110283
|
iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr15_-_18209672 | 1.07 |
ENSDART00000141508
ENSDART00000136280 |
btr16
|
bloodthirsty-related gene family, member 16 |
| chr21_-_17956739 | 1.06 |
ENSDART00000148154
|
stx2a
|
syntaxin 2a |
| chr6_+_112579 | 1.06 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr19_+_40350468 | 1.06 |
ENSDART00000087444
|
hepacam2
|
HEPACAM family member 2 |
| chr17_+_27456804 | 1.06 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr7_-_38659477 | 1.06 |
ENSDART00000138071
|
npsn
|
nephrosin |
| chr22_+_19522982 | 1.05 |
ENSDART00000192428
ENSDART00000190812 |
si:dkey-78l4.13
|
si:dkey-78l4.13 |
| chr19_-_3741602 | 1.05 |
ENSDART00000170301
|
btr22
|
bloodthirsty-related gene family, member 22 |
| chr16_-_41488023 | 1.04 |
ENSDART00000169312
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr20_+_2039518 | 1.04 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr21_+_28747236 | 1.04 |
ENSDART00000137874
|
zgc:100829
|
zgc:100829 |
| chr3_-_34561624 | 1.03 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr17_-_27382826 | 1.03 |
ENSDART00000186657
ENSDART00000155986 ENSDART00000191060 ENSDART00000077608 |
si:ch1073-358c10.1
|
si:ch1073-358c10.1 |
| chr11_+_141504 | 1.02 |
ENSDART00000086166
|
NCKAP1L
|
zgc:172352 |
| chr17_-_6202816 | 1.02 |
ENSDART00000151908
|
ptk2ba
|
protein tyrosine kinase 2 beta, a |
| chr3_-_40528333 | 1.02 |
ENSDART00000193047
|
actb2
|
actin, beta 2 |
| chr1_-_45347393 | 1.01 |
ENSDART00000173024
|
si:ch211-243a20.4
|
si:ch211-243a20.4 |
| chr9_+_24088062 | 1.01 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr17_-_29213710 | 1.01 |
ENSDART00000076481
|
ehd4
|
EH-domain containing 4 |
| chr18_+_13182528 | 1.01 |
ENSDART00000166298
|
zgc:56622
|
zgc:56622 |
| chr25_+_5068442 | 1.01 |
ENSDART00000097522
|
parvg
|
parvin, gamma |
| chr1_-_55942055 | 0.99 |
ENSDART00000142371
|
si:ch73-343l4.2
|
si:ch73-343l4.2 |
| chr1_-_18772241 | 0.99 |
ENSDART00000147645
ENSDART00000133194 ENSDART00000147525 ENSDART00000191493 |
tbc1d1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr15_-_36533322 | 0.98 |
ENSDART00000156466
ENSDART00000121755 |
si:dkey-262k9.4
|
si:dkey-262k9.4 |
| chr14_+_34501245 | 0.98 |
ENSDART00000131424
|
lcp2b
|
lymphocyte cytosolic protein 2b |
| chr9_-_33329700 | 0.98 |
ENSDART00000147265
ENSDART00000140039 |
rpl8
|
ribosomal protein L8 |
| chr17_+_53428092 | 0.98 |
ENSDART00000192509
|
srsf10b
|
serine/arginine-rich splicing factor 10b |
| chr3_-_23474416 | 0.98 |
ENSDART00000161672
|
si:dkey-225f5.5
|
si:dkey-225f5.5 |
| chr2_-_30182353 | 0.98 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr2_-_6482240 | 0.98 |
ENSDART00000132623
|
rgs13
|
regulator of G protein signaling 13 |
| chr14_+_3038473 | 0.97 |
ENSDART00000026021
ENSDART00000150000 |
cd74a
|
CD74 molecule, major histocompatibility complex, class II invariant chain a |
| chr21_-_31207522 | 0.97 |
ENSDART00000191637
|
zgc:152891
|
zgc:152891 |
| chr6_-_24143923 | 0.97 |
ENSDART00000157948
|
si:ch73-389b16.1
|
si:ch73-389b16.1 |
| chr20_+_19176959 | 0.96 |
ENSDART00000163276
ENSDART00000020099 |
blk
|
BLK proto-oncogene, Src family tyrosine kinase |
| chr6_-_14010554 | 0.96 |
ENSDART00000004656
|
zgc:92027
|
zgc:92027 |
| chr7_+_24573721 | 0.95 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr19_+_17386393 | 0.95 |
ENSDART00000034837
|
rpl15
|
ribosomal protein L15 |
| chr5_+_54938634 | 0.95 |
ENSDART00000163050
ENSDART00000145765 |
cd180
|
CD180 molecule |
| chr18_+_46162204 | 0.94 |
ENSDART00000113545
ENSDART00000147556 |
zgc:113340
|
zgc:113340 |
| chr12_+_10134366 | 0.94 |
ENSDART00000127343
|
smarcd2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr22_+_9806867 | 0.94 |
ENSDART00000137182
ENSDART00000106103 |
si:ch211-236g6.1
|
si:ch211-236g6.1 |
| chr7_-_57637779 | 0.94 |
ENSDART00000028017
|
mad2l1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr16_-_46619967 | 0.94 |
ENSDART00000158341
|
tmem176l.3a
|
transmembrane protein 176l.3a |
| chr23_+_17981127 | 0.93 |
ENSDART00000012571
ENSDART00000145200 |
chia.6
|
chitinase, acidic.6 |
| chr22_+_19528851 | 0.93 |
ENSDART00000145079
|
si:dkey-78l4.13
|
si:dkey-78l4.13 |
| chr21_+_33454147 | 0.93 |
ENSDART00000053208
|
rps14
|
ribosomal protein S14 |
| chr16_+_35395597 | 0.93 |
ENSDART00000158143
|
si:dkey-34d22.5
|
si:dkey-34d22.5 |
| chr17_-_21832119 | 0.91 |
ENSDART00000153539
ENSDART00000079008 |
si:ch211-208g24.8
|
si:ch211-208g24.8 |
| chr6_+_7977611 | 0.91 |
ENSDART00000112290
|
palm3
|
paralemmin 3 |
| chr1_-_46505310 | 0.91 |
ENSDART00000178072
|
si:busm1-105l16.2
|
si:busm1-105l16.2 |
| chr23_+_17980875 | 0.91 |
ENSDART00000163452
|
chia.6
|
chitinase, acidic.6 |
| chr15_-_33834577 | 0.90 |
ENSDART00000163354
|
mmp13b
|
matrix metallopeptidase 13b |
| chr6_+_28141135 | 0.90 |
ENSDART00000113405
ENSDART00000179795 |
inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr7_-_16596938 | 0.89 |
ENSDART00000134548
|
e2f8
|
E2F transcription factor 8 |
| chr20_-_34711528 | 0.89 |
ENSDART00000061555
|
si:ch211-63o20.7
|
si:ch211-63o20.7 |
| chr10_+_13209580 | 0.89 |
ENSDART00000000887
ENSDART00000136932 |
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr5_-_26879302 | 0.89 |
ENSDART00000098571
ENSDART00000139086 |
zgc:64051
|
zgc:64051 |
| chr21_+_28747069 | 0.89 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr23_+_36083529 | 0.88 |
ENSDART00000053295
ENSDART00000130260 |
hoxc10a
|
homeobox C10a |
| chr6_+_13046720 | 0.87 |
ENSDART00000165896
|
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr22_-_15577060 | 0.86 |
ENSDART00000176291
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr1_+_12394205 | 0.85 |
ENSDART00000138622
ENSDART00000136421 ENSDART00000139440 ENSDART00000184296 ENSDART00000008127 |
zgc:77739
|
zgc:77739 |
| chr15_+_5086338 | 0.85 |
ENSDART00000155402
|
mxf
|
myxovirus (influenza virus) resistance F |
| chr23_+_16948049 | 0.85 |
ENSDART00000139449
|
si:dkey-147f3.4
|
si:dkey-147f3.4 |
| chr16_+_46684855 | 0.85 |
ENSDART00000058325
|
lamtor2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr19_-_11431278 | 0.84 |
ENSDART00000109889
|
hsbp1l1
|
heat shock factor binding protein 1 like 1 |
| chr7_+_4795707 | 0.84 |
ENSDART00000142704
|
si:dkey-28d5.4
|
si:dkey-28d5.4 |
| chr8_-_32385989 | 0.84 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr4_-_13509946 | 0.84 |
ENSDART00000134720
|
ifng1-2
|
interferon, gamma 1-2 |
| chr6_+_52212927 | 0.83 |
ENSDART00000143458
|
ywhaba
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide a |
| chr5_-_24029228 | 0.83 |
ENSDART00000051546
|
rps6ka3a
|
ribosomal protein S6 kinase a, polypeptide 3a |
| chr16_-_33104944 | 0.83 |
ENSDART00000151943
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr12_+_19208338 | 0.83 |
ENSDART00000110415
|
sh3bp1
|
SH3-domain binding protein 1 |
| chr2_-_45510699 | 0.83 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr21_+_19062124 | 0.82 |
ENSDART00000134746
|
rpl17
|
ribosomal protein L17 |
| chr1_-_56213723 | 0.82 |
ENSDART00000142505
ENSDART00000137237 |
si:dkey-76b14.2
|
si:dkey-76b14.2 |
| chr2_-_13268556 | 0.82 |
ENSDART00000127693
ENSDART00000079757 |
vps4b
|
vacuolar protein sorting 4b homolog B (S. cerevisiae) |
| chr5_+_27421639 | 0.81 |
ENSDART00000146285
|
cyb561a3a
|
cytochrome b561 family, member A3a |
| chr16_-_46605160 | 0.81 |
ENSDART00000189070
ENSDART00000181194 |
tmem176l.3a
|
transmembrane protein 176l.3a |
| chr2_-_24068848 | 0.80 |
ENSDART00000145526
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr7_+_26545502 | 0.80 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of spi2+spic
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.6 | GO:0045627 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.6 | 1.9 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.6 | 1.7 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.5 | 2.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.5 | 1.6 | GO:0010525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.5 | 2.5 | GO:2000391 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.5 | 1.0 | GO:0002828 | regulation of type 2 immune response(GO:0002828) |
| 0.5 | 1.0 | GO:2000108 | positive regulation of leukocyte apoptotic process(GO:2000108) |
| 0.4 | 1.3 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.4 | 1.6 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.4 | 2.0 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.4 | 1.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 1.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.3 | 0.9 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.3 | 1.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.3 | 1.4 | GO:0046102 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.3 | 1.7 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.2 | 1.6 | GO:0071623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.2 | 0.7 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.2 | 0.6 | GO:0032466 | abscission(GO:0009838) negative regulation of cytokinesis(GO:0032466) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.2 | 1.5 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.2 | 1.2 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.2 | 1.8 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.2 | 2.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 2.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.6 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 1.5 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.5 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.1 | 0.8 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.4 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.1 | 2.5 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 0.4 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 5.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 0.6 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.6 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 1.5 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.1 | 1.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 0.7 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.7 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 1.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 3.9 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.9 | GO:0072422 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.1 | 0.3 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 0.9 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 1.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.3 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 0.5 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 0.4 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.1 | 1.0 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 0.4 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 0.4 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.8 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.4 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 1.7 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.1 | 0.2 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.1 | 2.8 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 1.2 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.1 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.4 | GO:0048940 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.1 | 0.4 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 2.5 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.1 | 0.5 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 1.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 1.0 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.3 | GO:0060043 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.1 | 1.4 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 0.4 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.1 | 2.5 | GO:0042113 | B cell activation(GO:0042113) |
| 0.1 | 2.1 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.3 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 1.0 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.4 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 6.7 | GO:0006956 | complement activation(GO:0006956) |
| 0.1 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.6 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 1.1 | GO:0039014 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.1 | 0.3 | GO:0052746 | inositol phosphorylation(GO:0052746) |
| 0.1 | 0.3 | GO:0071267 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 | 1.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.3 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.5 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 | 1.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.9 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.6 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.2 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.1 | 1.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 1.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.3 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.6 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.9 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.1 | 2.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 0.3 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 0.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.5 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.1 | 1.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:0032370 | intracellular cholesterol transport(GO:0032367) positive regulation of lipid transport(GO:0032370) |
| 0.0 | 0.8 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 2.3 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 1.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.4 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 4.7 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 2.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.2 | GO:0048855 | adenohypophysis morphogenesis(GO:0048855) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.4 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.2 | GO:0060343 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.0 | 0.4 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.4 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 1.2 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.3 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.8 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.7 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.5 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.3 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.4 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.9 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.8 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 1.5 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 1.6 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.4 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 2.7 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.7 | GO:0031577 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 1.0 | GO:0030098 | lymphocyte differentiation(GO:0030098) |
| 0.0 | 2.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.3 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 1.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.1 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.4 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.4 | GO:0051560 | calcium ion transmembrane import into mitochondrion(GO:0036444) mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.0 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.2 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 9.6 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.6 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0032945 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) negative regulation of leukocyte proliferation(GO:0070664) |
| 0.0 | 5.9 | GO:0032970 | regulation of actin filament-based process(GO:0032970) |
| 0.0 | 0.4 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.2 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.9 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.6 | GO:0009154 | purine ribonucleotide catabolic process(GO:0009154) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.5 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.4 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 6.5 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.9 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.2 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 1.2 | GO:0048565 | digestive tract development(GO:0048565) |
| 0.0 | 5.8 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.7 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.1 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.3 | 1.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 1.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 1.0 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 1.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 0.6 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 6.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.4 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 1.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.6 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.7 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.3 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 5.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 5.8 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.1 | 0.8 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.8 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 1.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 14.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.9 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 1.3 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 4.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 2.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 1.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 1.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.8 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.6 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 16.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.4 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 7.3 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.6 | 1.9 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.6 | 2.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.4 | 1.3 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.4 | 1.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.3 | 1.0 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.3 | 1.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 0.7 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.2 | 0.6 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.2 | 1.6 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.2 | 1.0 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 1.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 1.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 0.8 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.2 | 0.6 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.2 | 1.8 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 1.7 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 1.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 0.5 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 3.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 0.8 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 0.4 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.4 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.1 | 6.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.4 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.1 | 1.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.4 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.6 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 5.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.5 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.0 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 0.4 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.1 | 3.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.8 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.3 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.6 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.4 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.1 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.3 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 1.8 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.5 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 1.6 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.9 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 1.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 4.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 3.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.6 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.6 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 3.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 5.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 3.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.7 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.4 | GO:0008026 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 1.5 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 9.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 5.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0052813 | phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 3.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.6 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 8.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 2.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 1.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.1 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.0 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.6 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.1 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 0.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 2.0 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 0.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 5.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.9 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 1.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 0.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 1.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 1.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 1.9 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 2.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 1.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 2.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 2.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 1.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 5.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 5.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 2.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 2.8 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.3 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |