Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
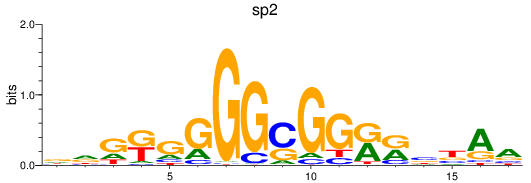
Results for sp2
Z-value: 0.65
Transcription factors associated with sp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sp2
|
ENSDARG00000076763 | sp2 transcription factor |
|
sp2
|
ENSDARG00000113443 | sp2 transcription factor |
|
sp2
|
ENSDARG00000115477 | sp2 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sp2 | dr11_v1_chr11_-_12008001_12008001 | 0.26 | 6.7e-01 | Click! |
Activity profile of sp2 motif
Sorted Z-values of sp2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_59015878 | 0.75 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr20_-_39219537 | 0.63 |
ENSDART00000005764
|
cyp39a1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr24_-_20599781 | 0.57 |
ENSDART00000179664
ENSDART00000141823 |
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr7_-_6592142 | 0.51 |
ENSDART00000160137
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr3_-_16227683 | 0.49 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr6_-_13408680 | 0.40 |
ENSDART00000151566
|
fmnl2b
|
formin-like 2b |
| chr2_-_44282796 | 0.39 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr8_+_23165749 | 0.37 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr4_+_19534833 | 0.36 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr1_+_54677173 | 0.36 |
ENSDART00000114705
|
gprc5bb
|
G protein-coupled receptor, class C, group 5, member Bb |
| chr2_-_11912347 | 0.35 |
ENSDART00000023851
|
abhd3
|
abhydrolase domain containing 3 |
| chr6_-_58828398 | 0.35 |
ENSDART00000090634
|
kif5ab
|
kinesin family member 5A, b |
| chr11_-_97817 | 0.33 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr11_-_32723851 | 0.32 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr10_+_1849874 | 0.32 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr3_-_36440705 | 0.30 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr8_-_4618653 | 0.29 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr6_-_58828113 | 0.29 |
ENSDART00000180934
|
kif5ab
|
kinesin family member 5A, b |
| chr13_+_16521898 | 0.29 |
ENSDART00000122557
|
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr2_+_24868010 | 0.29 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr5_+_26075230 | 0.29 |
ENSDART00000098473
|
klf9
|
Kruppel-like factor 9 |
| chr10_+_15777258 | 0.28 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr13_+_23677949 | 0.28 |
ENSDART00000144215
|
pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr10_-_641609 | 0.27 |
ENSDART00000041236
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr10_+_15777064 | 0.27 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr1_-_22861348 | 0.27 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr7_+_48460239 | 0.27 |
ENSDART00000052113
|
lingo1b
|
leucine rich repeat and Ig domain containing 1b |
| chr19_+_1097393 | 0.26 |
ENSDART00000168218
|
CABZ01111953.1
|
|
| chr8_+_36803415 | 0.26 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr9_+_42095220 | 0.25 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr24_+_41931585 | 0.24 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr5_-_38506981 | 0.23 |
ENSDART00000097822
|
atp1b2b
|
ATPase Na+/K+ transporting subunit beta 2b |
| chr20_+_52389858 | 0.23 |
ENSDART00000185863
ENSDART00000166651 |
arhgap39
|
Rho GTPase activating protein 39 |
| chr14_-_32503363 | 0.23 |
ENSDART00000034883
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr15_-_20731638 | 0.23 |
ENSDART00000170616
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr10_+_21650828 | 0.23 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr19_+_2279051 | 0.22 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr15_+_24388782 | 0.22 |
ENSDART00000191661
ENSDART00000179995 ENSDART00000111226 |
sez6b
|
seizure related 6 homolog b |
| chr1_-_55196103 | 0.22 |
ENSDART00000140153
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr24_-_28381404 | 0.22 |
ENSDART00000148406
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr6_-_35738836 | 0.22 |
ENSDART00000111642
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr23_+_44732863 | 0.21 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr18_-_2727764 | 0.21 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr24_-_30096666 | 0.21 |
ENSDART00000183285
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr1_+_44838706 | 0.20 |
ENSDART00000162779
ENSDART00000147357 |
kdm2aa
|
lysine (K)-specific demethylase 2Aa |
| chr9_-_24413008 | 0.20 |
ENSDART00000135897
|
tmeff2a
|
transmembrane protein with EGF-like and two follistatin-like domains 2a |
| chr3_-_16227490 | 0.20 |
ENSDART00000057159
ENSDART00000130611 ENSDART00000012835 |
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr11_-_35975026 | 0.20 |
ENSDART00000186219
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr10_+_37500234 | 0.20 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr3_-_5964557 | 0.20 |
ENSDART00000184738
|
BX284638.1
|
|
| chr11_-_44030962 | 0.20 |
ENSDART00000171910
|
FP016005.1
|
|
| chr11_-_41130239 | 0.20 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr18_+_21122818 | 0.20 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr11_+_19370717 | 0.19 |
ENSDART00000165906
|
prickle2b
|
prickle homolog 2b |
| chr2_+_37480669 | 0.19 |
ENSDART00000029801
|
sppl2
|
signal peptide peptidase-like 2 |
| chr11_-_1967438 | 0.19 |
ENSDART00000155844
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr22_+_12366516 | 0.19 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr4_-_22519516 | 0.19 |
ENSDART00000130409
ENSDART00000186258 ENSDART00000002851 ENSDART00000123801 |
kdm7aa
|
lysine (K)-specific demethylase 7Aa |
| chr20_+_37844035 | 0.19 |
ENSDART00000041397
|
flvcr1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr13_-_638485 | 0.18 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr1_+_39553040 | 0.18 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr9_-_202805 | 0.18 |
ENSDART00000182260
|
CABZ01067557.1
|
|
| chr2_+_24867534 | 0.18 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr12_+_19036380 | 0.18 |
ENSDART00000153086
ENSDART00000181060 |
kctd17
|
potassium channel tetramerization domain containing 17 |
| chr19_-_7450796 | 0.18 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr7_-_52876683 | 0.18 |
ENSDART00000172910
|
map1aa
|
microtubule-associated protein 1Aa |
| chr3_+_62126981 | 0.17 |
ENSDART00000060527
|
drc3
|
dynein regulatory complex subunit 3 |
| chr18_+_50907675 | 0.17 |
ENSDART00000159950
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr11_+_19370447 | 0.17 |
ENSDART00000186154
|
prickle2b
|
prickle homolog 2b |
| chr16_-_36798783 | 0.17 |
ENSDART00000145697
|
calb1
|
calbindin 1 |
| chr23_-_30960506 | 0.17 |
ENSDART00000142661
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr15_-_47956388 | 0.16 |
ENSDART00000116506
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr17_-_43287290 | 0.16 |
ENSDART00000156885
|
EML5
|
si:dkey-1f12.3 |
| chr9_+_13714379 | 0.16 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr18_+_27077853 | 0.16 |
ENSDART00000125326
ENSDART00000192660 ENSDART00000098334 |
ppp1r15b
|
protein phosphatase 1, regulatory subunit 15B |
| chr15_+_29393519 | 0.16 |
ENSDART00000193488
ENSDART00000112375 |
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr16_-_1757521 | 0.16 |
ENSDART00000124660
|
ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr21_-_40557281 | 0.16 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr1_-_633356 | 0.16 |
ENSDART00000171019
|
appa
|
amyloid beta (A4) precursor protein a |
| chr14_-_237130 | 0.16 |
ENSDART00000164988
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr16_+_1353894 | 0.16 |
ENSDART00000148426
|
celf3b
|
cugbp, Elav-like family member 3b |
| chr12_-_33893381 | 0.16 |
ENSDART00000153280
|
sema4gb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Gb |
| chr14_-_46113321 | 0.15 |
ENSDART00000169040
ENSDART00000161475 ENSDART00000124925 |
si:ch211-235e9.8
|
si:ch211-235e9.8 |
| chr23_+_2825940 | 0.15 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr21_+_21279159 | 0.15 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr6_+_27624023 | 0.15 |
ENSDART00000147789
|
slco2a1
|
solute carrier organic anion transporter family, member 2A1 |
| chr8_-_22739757 | 0.15 |
ENSDART00000182167
ENSDART00000171891 |
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr3_+_22578369 | 0.14 |
ENSDART00000187695
ENSDART00000182678 ENSDART00000112270 |
tanc2a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a |
| chr13_+_3819475 | 0.14 |
ENSDART00000139958
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr3_-_62380146 | 0.14 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr19_-_26769867 | 0.14 |
ENSDART00000043776
ENSDART00000159489 ENSDART00000138675 |
prrc2a
|
proline-rich coiled-coil 2A |
| chr15_-_14552101 | 0.14 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr19_-_26770083 | 0.14 |
ENSDART00000193811
ENSDART00000174455 |
prrc2a
|
proline-rich coiled-coil 2A |
| chr21_+_5589923 | 0.14 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr14_+_33264303 | 0.14 |
ENSDART00000130680
ENSDART00000075187 |
pdzd11
|
PDZ domain containing 11 |
| chr12_-_47899497 | 0.13 |
ENSDART00000162219
|
QRFPR
|
pyroglutamylated RFamide peptide receptor |
| chr1_-_55210619 | 0.13 |
ENSDART00000111671
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr17_+_51764310 | 0.13 |
ENSDART00000157171
|
si:ch211-168d23.3
|
si:ch211-168d23.3 |
| chr7_-_69636502 | 0.13 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr14_-_6225336 | 0.13 |
ENSDART00000111681
|
hdx
|
highly divergent homeobox |
| chr4_-_23839789 | 0.13 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr2_+_42592851 | 0.13 |
ENSDART00000135358
|
march11
|
membrane-associated ring finger (C3HC4) 11 |
| chr6_+_2271559 | 0.13 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr24_+_40473032 | 0.13 |
ENSDART00000084238
ENSDART00000178508 |
CABZ01076968.1
|
|
| chr10_+_39199547 | 0.13 |
ENSDART00000075943
|
ei24
|
etoposide induced 2.4 |
| chr3_-_3327573 | 0.12 |
ENSDART00000136319
ENSDART00000166667 |
tmem184bb
|
transmembrane protein 184bb |
| chr15_-_20731297 | 0.12 |
ENSDART00000114464
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr8_-_5847533 | 0.12 |
ENSDART00000192489
|
CABZ01102147.1
|
|
| chr18_-_40684756 | 0.12 |
ENSDART00000113799
ENSDART00000139042 |
si:ch211-132b12.7
|
si:ch211-132b12.7 |
| chr3_+_12322170 | 0.12 |
ENSDART00000161227
|
glis2b
|
GLIS family zinc finger 2b |
| chr17_-_40956035 | 0.12 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr17_+_11507131 | 0.11 |
ENSDART00000013170
|
kif26ba
|
kinesin family member 26Ba |
| chr20_+_36623807 | 0.11 |
ENSDART00000149171
ENSDART00000062895 |
srp9
|
signal recognition particle 9 |
| chr15_+_20543770 | 0.11 |
ENSDART00000092357
|
sgsm2
|
small G protein signaling modulator 2 |
| chr5_+_51443009 | 0.11 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr18_-_14836600 | 0.11 |
ENSDART00000045232
|
mtss1la
|
metastasis suppressor 1-like a |
| chr13_+_1100197 | 0.11 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr10_-_22831611 | 0.11 |
ENSDART00000160115
|
per1a
|
period circadian clock 1a |
| chr20_-_34868814 | 0.11 |
ENSDART00000153049
|
stmn4
|
stathmin-like 4 |
| chr19_-_15855427 | 0.11 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr22_+_465269 | 0.11 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr20_-_14781904 | 0.10 |
ENSDART00000187200
ENSDART00000179912 ENSDART00000160481 ENSDART00000026969 |
suco
|
SUN domain containing ossification factor |
| chr12_+_30109698 | 0.10 |
ENSDART00000042572
ENSDART00000153025 |
ablim1b
|
actin binding LIM protein 1b |
| chr17_+_27723490 | 0.10 |
ENSDART00000123588
ENSDART00000170462 ENSDART00000169708 |
qkia
|
QKI, KH domain containing, RNA binding a |
| chr19_+_2726819 | 0.10 |
ENSDART00000187122
ENSDART00000112414 |
rapgef5a
|
Rap guanine nucleotide exchange factor (GEF) 5a |
| chr18_-_46882649 | 0.10 |
ENSDART00000192056
|
gramd1bb
|
GRAM domain containing 1Bb |
| chr3_+_14388010 | 0.10 |
ENSDART00000171726
ENSDART00000165452 |
tmem56b
|
transmembrane protein 56b |
| chr21_-_11054876 | 0.10 |
ENSDART00000146576
|
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr3_+_35611625 | 0.10 |
ENSDART00000190995
|
traf7
|
TNF receptor-associated factor 7 |
| chr18_-_14836862 | 0.10 |
ENSDART00000124843
|
mtss1la
|
metastasis suppressor 1-like a |
| chr9_-_38156894 | 0.09 |
ENSDART00000134759
|
si:dkey-219c10.4
|
si:dkey-219c10.4 |
| chr11_+_7432533 | 0.09 |
ENSDART00000180977
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr12_-_42214 | 0.09 |
ENSDART00000045071
|
foxk2
|
forkhead box K2 |
| chr10_-_44924289 | 0.09 |
ENSDART00000171267
|
tuba7l
|
tubulin, alpha 7 like |
| chr14_+_1719367 | 0.09 |
ENSDART00000157696
|
trpc7b
|
transient receptor potential cation channel, subfamily C, member 7b |
| chr18_-_3166726 | 0.09 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr13_-_42724645 | 0.09 |
ENSDART00000046066
|
capn1a
|
calpain 1, (mu/I) large subunit a |
| chr15_-_9421481 | 0.09 |
ENSDART00000189045
ENSDART00000177158 |
sacs
|
sacsin molecular chaperone |
| chr19_+_935565 | 0.09 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr20_-_1268863 | 0.09 |
ENSDART00000109321
ENSDART00000027119 |
lats1
|
large tumor suppressor kinase 1 |
| chr3_+_51684963 | 0.09 |
ENSDART00000091180
ENSDART00000183711 ENSDART00000159493 |
baiap2a
|
BAI1-associated protein 2a |
| chr20_-_28931901 | 0.09 |
ENSDART00000153082
ENSDART00000046042 |
susd6
|
sushi domain containing 6 |
| chr5_-_14326959 | 0.09 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr11_-_36341189 | 0.09 |
ENSDART00000159752
|
sort1a
|
sortilin 1a |
| chr6_+_475264 | 0.09 |
ENSDART00000193615
|
LO017974.1
|
|
| chr21_-_11054605 | 0.09 |
ENSDART00000191378
ENSDART00000084061 |
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr14_+_24283915 | 0.08 |
ENSDART00000172868
|
klhl3
|
kelch-like family member 3 |
| chr14_-_45394704 | 0.08 |
ENSDART00000173078
ENSDART00000125970 |
si:ch211-168f7.5
|
si:ch211-168f7.5 |
| chr1_+_51329069 | 0.08 |
ENSDART00000187729
ENSDART00000193246 ENSDART00000186753 ENSDART00000190206 |
mast1a
|
microtubule associated serine/threonine kinase 1a |
| chr23_-_42093537 | 0.08 |
ENSDART00000122074
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr25_-_6389713 | 0.08 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr18_+_507835 | 0.08 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr15_-_43768776 | 0.08 |
ENSDART00000170398
|
grm5b
|
glutamate receptor, metabotropic 5b |
| chr8_-_4100365 | 0.08 |
ENSDART00000142846
|
cux2b
|
cut-like homeobox 2b |
| chr16_+_4839078 | 0.08 |
ENSDART00000150111
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr10_+_39200213 | 0.08 |
ENSDART00000153727
|
ei24
|
etoposide induced 2.4 |
| chr8_-_53490376 | 0.08 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr5_-_12743196 | 0.08 |
ENSDART00000188976
ENSDART00000137705 |
lztr1
|
leucine-zipper-like transcription regulator 1 |
| chr14_+_1687357 | 0.07 |
ENSDART00000174675
|
hrh2a
|
histamine receptor H2a |
| chr13_-_18069421 | 0.07 |
ENSDART00000146772
ENSDART00000134477 |
zfand4
|
zinc finger, AN1-type domain 4 |
| chr2_+_38881165 | 0.07 |
ENSDART00000141850
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr21_-_13972745 | 0.07 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr11_-_36341028 | 0.07 |
ENSDART00000146093
|
sort1a
|
sortilin 1a |
| chr5_-_13086616 | 0.07 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr16_+_4838808 | 0.07 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr22_+_567733 | 0.07 |
ENSDART00000171036
|
usp49
|
ubiquitin specific peptidase 49 |
| chr8_+_2656231 | 0.07 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr23_+_4890693 | 0.07 |
ENSDART00000023537
|
tnnc1a
|
troponin C type 1a (slow) |
| chr17_+_3379673 | 0.07 |
ENSDART00000176354
|
sntg2
|
syntrophin, gamma 2 |
| chr15_-_11955485 | 0.06 |
ENSDART00000160286
|
si:dkey-202l22.3
|
si:dkey-202l22.3 |
| chr5_-_10002260 | 0.06 |
ENSDART00000141831
|
si:ch73-266o15.4
|
si:ch73-266o15.4 |
| chr7_+_38249858 | 0.06 |
ENSDART00000150158
|
si:dkey-78a14.4
|
si:dkey-78a14.4 |
| chr12_+_14149686 | 0.06 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr17_-_37474689 | 0.06 |
ENSDART00000103980
|
crip2
|
cysteine-rich protein 2 |
| chr6_+_15373153 | 0.06 |
ENSDART00000155865
|
tmtops2a
|
teleost multiple tissue opsin 2a |
| chr10_+_44924684 | 0.06 |
ENSDART00000181360
ENSDART00000170418 ENSDART00000170327 |
sec14l7
|
SEC14-like lipid binding 7 |
| chr7_-_48396193 | 0.06 |
ENSDART00000083555
|
sin3ab
|
SIN3 transcription regulator family member Ab |
| chr15_-_25435085 | 0.06 |
ENSDART00000112079
|
tlcd2
|
TLC domain containing 2 |
| chr23_+_41200854 | 0.06 |
ENSDART00000109567
|
nhsa
|
Nance-Horan syndrome a (congenital cataracts and dental anomalies) |
| chr19_-_2029777 | 0.05 |
ENSDART00000128639
|
CABZ01071939.1
|
|
| chr24_+_10039165 | 0.05 |
ENSDART00000144186
|
pou6f2
|
POU class 6 homeobox 2 |
| chr7_+_38962207 | 0.05 |
ENSDART00000173565
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr15_-_6976851 | 0.05 |
ENSDART00000158474
ENSDART00000168943 ENSDART00000169944 |
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr9_+_23825440 | 0.05 |
ENSDART00000138470
|
ints6
|
integrator complex subunit 6 |
| chr3_+_11548516 | 0.05 |
ENSDART00000059117
|
mmd
|
monocyte to macrophage differentiation-associated |
| chr19_+_13994563 | 0.05 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr1_+_13087797 | 0.05 |
ENSDART00000170314
|
si:dkey-5n7.2
|
si:dkey-5n7.2 |
| chr12_-_14211293 | 0.05 |
ENSDART00000158399
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr23_-_18982499 | 0.05 |
ENSDART00000012507
|
bcl2l1
|
bcl2-like 1 |
| chr18_+_507618 | 0.05 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr19_+_26340736 | 0.05 |
ENSDART00000013497
|
mylipa
|
myosin regulatory light chain interacting protein a |
| chr2_+_25657958 | 0.05 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr25_-_5963535 | 0.04 |
ENSDART00000155751
|
nuak1b
|
NUAK family, SNF1-like kinase, 1b |
| chr20_-_16156419 | 0.04 |
ENSDART00000037420
|
ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr24_+_28935200 | 0.04 |
ENSDART00000017427
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr17_-_8268406 | 0.04 |
ENSDART00000149873
ENSDART00000064668 ENSDART00000148403 |
ahi1
|
Abelson helper integration site 1 |
| chr20_-_53981626 | 0.04 |
ENSDART00000023550
|
hsp90aa1.2
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 |
| chr7_-_5396154 | 0.04 |
ENSDART00000172980
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr19_-_31802296 | 0.04 |
ENSDART00000103640
|
hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr20_-_43786515 | 0.04 |
ENSDART00000004601
|
laptm4a
|
lysosomal protein transmembrane 4 alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of sp2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 0.4 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.6 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.1 | 0.6 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.3 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.1 | 0.5 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.4 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 0.2 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 0.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.1 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.0 | 0.2 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.4 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.2 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0090199 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.1 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.3 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.2 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.3 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.0 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.2 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 0.2 | GO:0097178 | ruffle assembly(GO:0097178) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.2 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.4 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.0 | GO:0030626 | U12 snRNA binding(GO:0030626) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |