Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
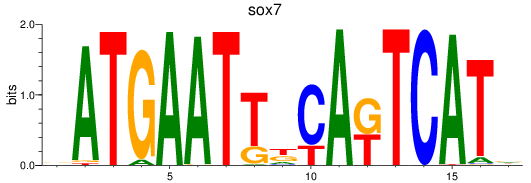
Results for sox7
Z-value: 0.54
Transcription factors associated with sox7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox7
|
ENSDARG00000030125 | SRY-box transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox7 | dr11_v1_chr20_+_19066858_19066858 | 0.74 | 1.5e-01 | Click! |
Activity profile of sox7 motif
Sorted Z-values of sox7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_29191152 | 0.54 |
ENSDART00000132702
|
pvalb7
|
parvalbumin 7 |
| chr22_-_29204960 | 0.51 |
ENSDART00000131386
|
pvalb7
|
parvalbumin 7 |
| chr15_-_23647078 | 0.50 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr2_-_24996441 | 0.39 |
ENSDART00000144795
|
slc35g2a
|
solute carrier family 35, member G2a |
| chr22_-_29205327 | 0.37 |
ENSDART00000183161
ENSDART00000189515 |
pvalb7
|
parvalbumin 7 |
| chr20_+_50956369 | 0.31 |
ENSDART00000170854
|
gphnb
|
gephyrin b |
| chr16_+_10777116 | 0.27 |
ENSDART00000190902
|
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr25_-_22639133 | 0.25 |
ENSDART00000073583
|
islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr16_+_46111849 | 0.24 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr1_-_470812 | 0.23 |
ENSDART00000192527
|
zgc:92518
|
zgc:92518 |
| chr8_+_23174137 | 0.23 |
ENSDART00000189470
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr20_-_25709247 | 0.21 |
ENSDART00000146711
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr13_+_24829996 | 0.21 |
ENSDART00000147194
|
si:ch211-223p8.8
|
si:ch211-223p8.8 |
| chr22_-_12862415 | 0.18 |
ENSDART00000145156
ENSDART00000137280 |
glsa
|
glutaminase a |
| chr17_-_26537928 | 0.18 |
ENSDART00000155692
ENSDART00000122366 |
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr14_+_6546516 | 0.16 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr11_+_10541258 | 0.16 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr17_-_22010668 | 0.16 |
ENSDART00000031998
|
slc22a7b.1
|
solute carrier family 22 (organic anion transporter), member 7b, tandem duplicate 1 |
| chr10_-_7913591 | 0.16 |
ENSDART00000139661
|
slc35e4
|
solute carrier family 35, member E4 |
| chr15_-_21692630 | 0.15 |
ENSDART00000039865
|
sdhdb
|
succinate dehydrogenase complex, subunit D, integral membrane protein b |
| chr17_+_15534815 | 0.14 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr12_+_26621906 | 0.14 |
ENSDART00000158440
ENSDART00000046959 |
arhgap12b
|
Rho GTPase activating protein 12b |
| chr14_-_30490465 | 0.14 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr11_-_35970966 | 0.14 |
ENSDART00000188805
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr14_-_6365223 | 0.13 |
ENSDART00000167446
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr18_-_39200557 | 0.13 |
ENSDART00000132367
ENSDART00000183672 |
si:ch211-235f12.2
mapk6
|
si:ch211-235f12.2 mitogen-activated protein kinase 6 |
| chr19_-_42462491 | 0.13 |
ENSDART00000131715
|
psmb4
|
proteasome subunit beta 4 |
| chr16_+_27345383 | 0.13 |
ENSDART00000078250
ENSDART00000162857 |
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr10_-_23379146 | 0.12 |
ENSDART00000056369
|
cadm2a
|
cell adhesion molecule 2a |
| chr25_-_7999756 | 0.11 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr5_-_69923241 | 0.10 |
ENSDART00000187389
|
fktn
|
fukutin |
| chr5_+_34982318 | 0.10 |
ENSDART00000160504
|
ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr22_-_26558166 | 0.10 |
ENSDART00000111125
|
glis2a
|
GLIS family zinc finger 2a |
| chr21_+_31811202 | 0.09 |
ENSDART00000189846
|
si:ch211-12m10.1
|
si:ch211-12m10.1 |
| chr19_-_11031145 | 0.09 |
ENSDART00000151375
ENSDART00000027598 ENSDART00000137865 ENSDART00000188025 |
tpm3
|
tropomyosin 3 |
| chr12_+_30109698 | 0.09 |
ENSDART00000042572
ENSDART00000153025 |
ablim1b
|
actin binding LIM protein 1b |
| chr9_-_23033818 | 0.09 |
ENSDART00000022392
|
rnd3b
|
Rho family GTPase 3b |
| chr12_+_4114401 | 0.09 |
ENSDART00000025912
|
si:dkey-32n7.4
|
si:dkey-32n7.4 |
| chr19_+_5480327 | 0.08 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr1_+_36674584 | 0.08 |
ENSDART00000186772
ENSDART00000192274 |
ednraa
|
endothelin receptor type Aa |
| chr11_+_44356707 | 0.08 |
ENSDART00000165219
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr11_-_3613558 | 0.08 |
ENSDART00000163578
|
dnajc16l
|
DnaJ (Hsp40) homolog, subfamily C, member 16 like |
| chr11_+_44356504 | 0.07 |
ENSDART00000160678
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr8_+_35664152 | 0.07 |
ENSDART00000144520
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr2_-_16986894 | 0.06 |
ENSDART00000145720
|
clcn2a
|
chloride channel, voltage-sensitive 2a |
| chr1_+_6206950 | 0.06 |
ENSDART00000146473
|
retreg2
|
reticulophagy regulator family member 2 |
| chr3_+_52737565 | 0.06 |
ENSDART00000108639
|
gmip
|
GEM interacting protein |
| chr24_-_20652473 | 0.05 |
ENSDART00000179984
|
nktr
|
natural killer cell triggering receptor |
| chr5_-_67911111 | 0.05 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr7_+_41421998 | 0.05 |
ENSDART00000083967
|
chpf2
|
chondroitin polymerizing factor 2 |
| chr20_-_4766645 | 0.05 |
ENSDART00000147071
ENSDART00000152398 |
ak7a
|
adenylate kinase 7a |
| chr22_+_2239254 | 0.04 |
ENSDART00000131396
ENSDART00000135320 |
znf1144
|
zinc finger protein 1144 |
| chr15_-_35252522 | 0.04 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr12_-_4540564 | 0.04 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr7_-_35083184 | 0.04 |
ENSDART00000100253
ENSDART00000135250 ENSDART00000173511 |
agrp
|
agouti related neuropeptide |
| chr1_-_49649122 | 0.03 |
ENSDART00000134399
|
slkb
|
STE20-like kinase b |
| chr3_-_51142942 | 0.03 |
ENSDART00000160789
|
si:ch211-148f13.1
|
si:ch211-148f13.1 |
| chr18_+_15876385 | 0.03 |
ENSDART00000142527
|
eea1
|
early endosome antigen 1 |
| chr13_+_10799380 | 0.03 |
ENSDART00000126427
|
lrpprc
|
leucine-rich pentatricopeptide repeat containing |
| chr9_+_38369872 | 0.02 |
ENSDART00000193914
|
plcd4b
|
phospholipase C, delta 4b |
| chr19_+_10860485 | 0.02 |
ENSDART00000169962
|
si:ch73-347e22.4
|
si:ch73-347e22.4 |
| chr24_+_35787629 | 0.02 |
ENSDART00000136721
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr16_+_40024883 | 0.02 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr8_-_20291922 | 0.02 |
ENSDART00000148304
|
myo1f
|
myosin IF |
| chr23_+_21261313 | 0.01 |
ENSDART00000104268
ENSDART00000159046 |
emc1
|
ER membrane protein complex subunit 1 |
| chr4_-_71311438 | 0.01 |
ENSDART00000168021
|
si:dkeyp-123a12.2
|
si:dkeyp-123a12.2 |
| chr7_-_4107423 | 0.01 |
ENSDART00000172971
|
zgc:55733
|
zgc:55733 |
| chr5_-_3960161 | 0.01 |
ENSDART00000111453
|
myo19
|
myosin XIX |
| chr8_+_2787042 | 0.00 |
ENSDART00000155275
|
sh3glb2a
|
SH3-domain GRB2-like endophilin B2a |
| chr14_+_1124409 | 0.00 |
ENSDART00000190043
ENSDART00000106708 ENSDART00000160246 |
sec24b
|
SEC24 homolog B, COPII coat complex component |
| chr22_-_26852516 | 0.00 |
ENSDART00000005829
|
gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr22_+_29204885 | 0.00 |
ENSDART00000133409
ENSDART00000059820 |
ncf4
|
neutrophil cytosolic factor 4 |
| chr13_+_43650632 | 0.00 |
ENSDART00000141024
|
zfyve21
|
zinc finger, FYVE domain containing 21 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0018315 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.5 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.2 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.0 | 0.3 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 0.2 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.5 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |