Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
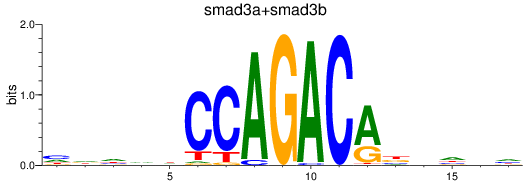
Results for smad3a+smad3b
Z-value: 1.90
Transcription factors associated with smad3a+smad3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
smad3b
|
ENSDARG00000010207 | SMAD family member 3b |
|
smad3a
|
ENSDARG00000036096 | SMAD family member 3a |
|
smad3a
|
ENSDARG00000117146 | SMAD family member 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| smad3a | dr11_v1_chr7_-_34149263_34149263 | -0.82 | 8.6e-02 | Click! |
| smad3b | dr11_v1_chr18_+_19772874_19772874 | -0.57 | 3.2e-01 | Click! |
Activity profile of smad3a+smad3b motif
Sorted Z-values of smad3a+smad3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_59073203 | 2.32 |
ENSDART00000149937
ENSDART00000162201 |
MFAP4 (1 of many)
zgc:173915
|
si:zfos-2330d3.3 zgc:173915 |
| chr22_+_661711 | 1.80 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr22_+_661505 | 1.75 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr5_+_22459087 | 1.58 |
ENSDART00000134781
|
BX546499.1
|
|
| chr19_-_5332784 | 1.55 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr1_-_59104145 | 1.48 |
ENSDART00000132495
ENSDART00000152457 |
MFAP4 (1 of many)
si:zfos-2330d3.7
|
si:zfos-2330d3.1 si:zfos-2330d3.7 |
| chr23_+_44614056 | 1.47 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr14_+_38786298 | 1.39 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr18_+_13164325 | 1.34 |
ENSDART00000189057
|
tat
|
tyrosine aminotransferase |
| chr1_+_59073436 | 1.34 |
ENSDART00000161642
|
MFAP4 (1 of many)
|
si:zfos-2330d3.3 |
| chr2_+_32796873 | 1.32 |
ENSDART00000077511
|
ccr9a
|
chemokine (C-C motif) receptor 9a |
| chr4_-_11577253 | 1.32 |
ENSDART00000144452
|
net1
|
neuroepithelial cell transforming 1 |
| chr10_+_17088261 | 1.29 |
ENSDART00000132103
|
si:dkey-106l3.7
|
si:dkey-106l3.7 |
| chr19_+_4068134 | 1.18 |
ENSDART00000158285
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr13_+_2830574 | 1.16 |
ENSDART00000138143
|
si:ch211-233m11.1
|
si:ch211-233m11.1 |
| chr6_+_49926115 | 1.15 |
ENSDART00000018523
|
ahcy
|
adenosylhomocysteinase |
| chr2_-_7666021 | 1.14 |
ENSDART00000180007
|
CABZ01021592.1
|
|
| chr3_-_48980319 | 1.10 |
ENSDART00000165319
|
ftr42
|
finTRIM family, member 42 |
| chr7_+_21180747 | 1.08 |
ENSDART00000185543
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr12_+_46386983 | 1.05 |
ENSDART00000183982
|
BX005305.3
|
Danio rerio legumain (LOC100005356), mRNA. |
| chr16_-_17699111 | 1.02 |
ENSDART00000108581
|
si:dkey-17m8.1
|
si:dkey-17m8.1 |
| chr19_+_4062101 | 1.00 |
ENSDART00000166773
|
btr25
|
bloodthirsty-related gene family, member 25 |
| chr25_+_35065802 | 0.97 |
ENSDART00000182372
|
zgc:165555
|
zgc:165555 |
| chr1_-_43920371 | 0.95 |
ENSDART00000109283
|
scpp7
|
secretory calcium-binding phosphoprotein 7 |
| chr3_-_4591643 | 0.94 |
ENSDART00000138144
|
ftr50
|
finTRIM family, member 50 |
| chr1_-_43920576 | 0.90 |
ENSDART00000191914
|
scpp7
|
secretory calcium-binding phosphoprotein 7 |
| chr18_+_20560442 | 0.89 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr5_-_24124118 | 0.89 |
ENSDART00000051550
|
capga
|
capping protein (actin filament), gelsolin-like a |
| chr13_+_17694845 | 0.87 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr15_+_46344655 | 0.86 |
ENSDART00000155893
|
si:ch1073-340i21.2
|
si:ch1073-340i21.2 |
| chr21_+_26071874 | 0.85 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr4_-_18954001 | 0.84 |
ENSDART00000144814
|
si:dkey-31f5.8
|
si:dkey-31f5.8 |
| chr19_+_627899 | 0.81 |
ENSDART00000148508
|
tert
|
telomerase reverse transcriptase |
| chr4_+_28997595 | 0.81 |
ENSDART00000133357
|
si:dkey-13e3.1
|
si:dkey-13e3.1 |
| chr1_-_52494122 | 0.80 |
ENSDART00000131407
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr18_-_26715655 | 0.78 |
ENSDART00000181497
|
malt3
|
MALT paracaspase 3 |
| chr8_-_38201415 | 0.78 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr7_+_2228276 | 0.77 |
ENSDART00000064294
|
si:dkey-187j14.4
|
si:dkey-187j14.4 |
| chr22_-_10591876 | 0.76 |
ENSDART00000105846
|
si:dkey-42i9.8
|
si:dkey-42i9.8 |
| chr19_+_4061699 | 0.74 |
ENSDART00000158309
ENSDART00000166512 |
btr25
btr26
|
bloodthirsty-related gene family, member 25 bloodthirsty-related gene family, member 26 |
| chr21_-_32036597 | 0.74 |
ENSDART00000114964
|
zgc:165573
|
zgc:165573 |
| chr22_+_14836291 | 0.74 |
ENSDART00000122740
|
gtpbp1l
|
GTP binding protein 1, like |
| chr15_-_25556074 | 0.72 |
ENSDART00000124677
|
mmp20a
|
matrix metallopeptidase 20a (enamelysin) |
| chr15_+_38007237 | 0.71 |
ENSDART00000182271
|
CR944667.2
|
|
| chr15_+_47525073 | 0.70 |
ENSDART00000067583
|
sidt2
|
SID1 transmembrane family, member 2 |
| chr10_-_36214582 | 0.70 |
ENSDART00000166471
|
or109-11
|
odorant receptor, family D, subfamily 109, member 11 |
| chr3_-_27784457 | 0.68 |
ENSDART00000019098
|
dnase1
|
deoxyribonuclease I |
| chr6_-_8498908 | 0.68 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr2_-_20599315 | 0.68 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr7_+_5959203 | 0.68 |
ENSDART00000110813
|
zgc:165555
|
zgc:165555 |
| chr4_+_15944245 | 0.67 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr7_+_4911404 | 0.67 |
ENSDART00000137385
|
si:dkey-28d5.11
|
si:dkey-28d5.11 |
| chr3_+_52753416 | 0.66 |
ENSDART00000171185
|
gmip
|
GEM interacting protein |
| chr24_-_6038025 | 0.66 |
ENSDART00000077819
ENSDART00000139216 |
ftr61
|
finTRIM family, member 61 |
| chr19_-_3754327 | 0.65 |
ENSDART00000168463
|
btr21
|
bloodthirsty-related gene family, member 21 |
| chr1_-_59130383 | 0.64 |
ENSDART00000171552
|
FP015850.1
|
|
| chr19_+_4054036 | 0.64 |
ENSDART00000171802
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr25_-_29415369 | 0.64 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr19_+_7043634 | 0.63 |
ENSDART00000133954
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr24_+_39990695 | 0.63 |
ENSDART00000040281
|
BX323854.1
|
|
| chr6_-_8498676 | 0.63 |
ENSDART00000148627
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr25_-_35126024 | 0.62 |
ENSDART00000185109
|
HIST1H4I
|
zgc:165555 |
| chr4_+_50401133 | 0.62 |
ENSDART00000150644
|
si:dkey-156k2.7
|
si:dkey-156k2.7 |
| chr12_-_21681509 | 0.61 |
ENSDART00000112726
|
eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr13_-_10945288 | 0.61 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr7_+_66822229 | 0.61 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr3_-_4501026 | 0.61 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr15_-_25556227 | 0.60 |
ENSDART00000156445
|
mmp20a
|
matrix metallopeptidase 20a (enamelysin) |
| chr25_+_16194979 | 0.60 |
ENSDART00000185592
ENSDART00000158582 ENSDART00000161109 ENSDART00000139013 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr5_+_38685089 | 0.60 |
ENSDART00000139743
|
si:dkey-58f10.10
|
si:dkey-58f10.10 |
| chr3_+_13603272 | 0.59 |
ENSDART00000185084
|
hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr12_-_44016898 | 0.59 |
ENSDART00000175304
|
si:dkey-201i2.4
|
si:dkey-201i2.4 |
| chr22_+_19478140 | 0.58 |
ENSDART00000135291
ENSDART00000136576 |
si:dkey-78l4.8
|
si:dkey-78l4.8 |
| chr16_-_53259409 | 0.58 |
ENSDART00000157080
|
si:ch211-269k10.4
|
si:ch211-269k10.4 |
| chr7_-_69121896 | 0.57 |
ENSDART00000130227
|
crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr14_-_32876280 | 0.57 |
ENSDART00000173168
|
si:rp71-46j2.7
|
si:rp71-46j2.7 |
| chr25_-_16552135 | 0.57 |
ENSDART00000125836
|
si:ch211-266k8.4
|
si:ch211-266k8.4 |
| chr2_+_42236118 | 0.56 |
ENSDART00000114162
ENSDART00000141199 |
ftr02
|
finTRIM family, member 2 |
| chr15_-_41486793 | 0.56 |
ENSDART00000138525
|
si:ch211-187g4.1
|
si:ch211-187g4.1 |
| chr13_+_10945337 | 0.56 |
ENSDART00000091845
|
abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr1_-_59139599 | 0.55 |
ENSDART00000152233
|
si:ch1073-110a20.3
|
si:ch1073-110a20.3 |
| chr10_-_24689725 | 0.55 |
ENSDART00000079566
|
si:ch211-287a12.9
|
si:ch211-287a12.9 |
| chr25_+_36315127 | 0.55 |
ENSDART00000191984
|
zgc:165555
|
zgc:165555 |
| chr7_+_24523017 | 0.55 |
ENSDART00000077047
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr9_+_29616854 | 0.55 |
ENSDART00000033902
ENSDART00000143493 |
phf11
|
PHD finger protein 11 |
| chr5_+_62723233 | 0.55 |
ENSDART00000183718
|
nanos2
|
nanos homolog 2 |
| chr24_-_33291784 | 0.54 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr2_-_59265521 | 0.54 |
ENSDART00000146341
ENSDART00000097799 |
ftr33
|
finTRIM family, member 33 |
| chr6_-_43283122 | 0.53 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr22_-_17499513 | 0.52 |
ENSDART00000105460
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr3_-_2091029 | 0.52 |
ENSDART00000141464
|
si:dkey-88j15.4
|
si:dkey-88j15.4 |
| chr14_-_7128980 | 0.51 |
ENSDART00000171311
|
si:ch73-43g23.1
|
si:ch73-43g23.1 |
| chr5_+_38752287 | 0.51 |
ENSDART00000133571
|
cxcl11.8
|
chemokine (C-X-C motif) ligand 11, duplicate 8 |
| chr15_-_37834433 | 0.50 |
ENSDART00000189748
|
si:dkey-238d18.3
|
si:dkey-238d18.3 |
| chr3_+_34120191 | 0.50 |
ENSDART00000020017
ENSDART00000151700 |
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr23_+_10352921 | 0.50 |
ENSDART00000081193
|
KRT18 (1 of many)
|
si:ch211-133j6.3 |
| chr10_-_24416471 | 0.50 |
ENSDART00000135985
|
hephl1b
|
hephaestin-like 1b |
| chr5_+_38673168 | 0.49 |
ENSDART00000135267
|
si:dkey-58f10.12
|
si:dkey-58f10.12 |
| chr20_-_52199296 | 0.48 |
ENSDART00000131806
ENSDART00000143668 |
hlx1
|
H2.0-like homeo box 1 (Drosophila) |
| chr4_+_35576329 | 0.48 |
ENSDART00000171191
ENSDART00000161611 |
si:dkeyp-4c4.2
|
si:dkeyp-4c4.2 |
| chr16_-_32204470 | 0.48 |
ENSDART00000143928
|
rwdd1
|
RWD domain containing 1 |
| chr20_-_2619316 | 0.47 |
ENSDART00000185777
|
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr17_-_27273296 | 0.46 |
ENSDART00000077087
|
id3
|
inhibitor of DNA binding 3 |
| chr17_+_14886828 | 0.45 |
ENSDART00000010507
ENSDART00000131052 |
ptger2a
|
prostaglandin E receptor 2a (subtype EP2) |
| chr23_-_19230627 | 0.45 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr25_+_36345867 | 0.44 |
ENSDART00000186527
|
zgc:165555
|
zgc:165555 |
| chr16_+_15114645 | 0.44 |
ENSDART00000158483
|
mtbp
|
MDM2 binding protein |
| chr14_-_40797117 | 0.44 |
ENSDART00000122369
|
elf1
|
E74-like ETS transcription factor 1 |
| chr18_-_8885792 | 0.43 |
ENSDART00000143619
|
si:dkey-95h12.1
|
si:dkey-95h12.1 |
| chr7_-_24181159 | 0.43 |
ENSDART00000181206
|
zgc:153151
|
zgc:153151 |
| chr25_+_35056619 | 0.42 |
ENSDART00000154389
|
HIST1H4J
|
histone cluster 1 H4 family member j |
| chr6_+_28051978 | 0.41 |
ENSDART00000143218
|
si:ch73-194h10.2
|
si:ch73-194h10.2 |
| chr2_+_25560556 | 0.41 |
ENSDART00000133623
|
pld1a
|
phospholipase D1a |
| chr7_-_51528661 | 0.40 |
ENSDART00000174263
|
nhsl2
|
NHS-like 2 |
| chr21_-_40173821 | 0.40 |
ENSDART00000180667
|
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr8_-_18203274 | 0.40 |
ENSDART00000134078
ENSDART00000180235 ENSDART00000080006 ENSDART00000125418 ENSDART00000142114 |
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr21_+_2203316 | 0.40 |
ENSDART00000185659
|
BX510922.4
|
|
| chr16_+_19013257 | 0.40 |
ENSDART00000144102
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr20_+_46213553 | 0.39 |
ENSDART00000100532
|
stx7l
|
syntaxin 7-like |
| chr7_-_3721671 | 0.38 |
ENSDART00000062732
|
si:ch211-282j17.8
|
si:ch211-282j17.8 |
| chr22_-_35385810 | 0.38 |
ENSDART00000111824
|
HTR3C
|
5-hydroxytryptamine receptor 3C |
| chr1_-_57129179 | 0.37 |
ENSDART00000157226
ENSDART00000152469 |
si:ch73-94k4.2
|
si:ch73-94k4.2 |
| chr14_-_28052474 | 0.37 |
ENSDART00000172948
ENSDART00000135337 |
TSC22D3 (1 of many)
zgc:64189
|
si:ch211-220e11.3 zgc:64189 |
| chr22_+_36914636 | 0.37 |
ENSDART00000150948
|
pimr205
|
Pim proto-oncogene, serine/threonine kinase, related 205 |
| chr12_+_16949391 | 0.37 |
ENSDART00000152635
|
zgc:174153
|
zgc:174153 |
| chr4_-_77636185 | 0.36 |
ENSDART00000152915
ENSDART00000131964 |
si:dkey-61p9.9
|
si:dkey-61p9.9 |
| chr12_-_44122412 | 0.35 |
ENSDART00000169094
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr17_-_45386546 | 0.35 |
ENSDART00000182647
|
tmem206
|
transmembrane protein 206 |
| chr12_-_44010532 | 0.35 |
ENSDART00000183875
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr6_-_39649504 | 0.35 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr18_-_22696170 | 0.34 |
ENSDART00000147183
|
si:dkey-11d18.4
|
si:dkey-11d18.4 |
| chr4_+_47736069 | 0.33 |
ENSDART00000193329
|
si:ch211-196f19.1
|
si:ch211-196f19.1 |
| chr6_-_39158953 | 0.33 |
ENSDART00000050682
|
stat2
|
signal transducer and activator of transcription 2 |
| chr2_-_59376399 | 0.33 |
ENSDART00000137134
|
ftr38
|
finTRIM family, member 38 |
| chr3_+_52275691 | 0.33 |
ENSDART00000190109
|
BX511132.2
|
|
| chr22_+_1526040 | 0.33 |
ENSDART00000164089
ENSDART00000159126 |
si:ch211-255f4.6
|
si:ch211-255f4.6 |
| chr12_+_4573696 | 0.32 |
ENSDART00000152534
|
si:dkey-94f20.4
|
si:dkey-94f20.4 |
| chr4_-_71267461 | 0.32 |
ENSDART00000161219
ENSDART00000151838 |
si:dkeyp-51g9.4
|
si:dkeyp-51g9.4 |
| chr12_-_44151296 | 0.32 |
ENSDART00000168734
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr8_-_3346692 | 0.31 |
ENSDART00000057874
|
FUT9 (1 of many)
|
zgc:103510 |
| chr10_+_23441488 | 0.31 |
ENSDART00000100783
|
si:ch73-122g19.1
|
si:ch73-122g19.1 |
| chr11_+_30310170 | 0.31 |
ENSDART00000127797
|
ugt1b3
|
UDP glucuronosyltransferase 1 family, polypeptide B3 |
| chr3_-_62417677 | 0.31 |
ENSDART00000101888
|
sstr2b
|
somatostatin receptor 2b |
| chr20_-_54377933 | 0.30 |
ENSDART00000182664
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr7_+_29890292 | 0.30 |
ENSDART00000170403
ENSDART00000168600 |
tln2a
|
talin 2a |
| chr18_-_22696357 | 0.29 |
ENSDART00000100955
|
si:dkey-11d18.4
|
si:dkey-11d18.4 |
| chr1_+_47178529 | 0.29 |
ENSDART00000158432
ENSDART00000074450 ENSDART00000137448 |
morc3b
|
MORC family CW-type zinc finger 3b |
| chr5_+_17780475 | 0.29 |
ENSDART00000110783
ENSDART00000115227 |
chfr
|
checkpoint with forkhead and ring finger domains, E3 ubiquitin protein ligase |
| chr4_-_73227710 | 0.29 |
ENSDART00000193165
|
LO018260.3
|
|
| chr22_-_6801876 | 0.29 |
ENSDART00000135726
|
si:ch1073-188e1.1
|
si:ch1073-188e1.1 |
| chr3_-_33029426 | 0.29 |
ENSDART00000181287
|
CT583642.1
|
|
| chr19_-_24218942 | 0.29 |
ENSDART00000189198
|
BX547993.2
|
|
| chr16_+_25259313 | 0.28 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr12_-_17479078 | 0.28 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr8_-_13541514 | 0.28 |
ENSDART00000063834
|
zgc:86586
|
zgc:86586 |
| chr8_-_3336273 | 0.27 |
ENSDART00000081160
|
zgc:173737
|
zgc:173737 |
| chr4_-_25812329 | 0.27 |
ENSDART00000146658
|
tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr18_-_31227615 | 0.26 |
ENSDART00000003210
|
mc1r
|
melanocortin 1 receptor |
| chr17_-_51679784 | 0.26 |
ENSDART00000103374
|
atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr2_-_55337585 | 0.26 |
ENSDART00000177924
|
tpm4b
|
tropomyosin 4b |
| chr10_+_36345176 | 0.25 |
ENSDART00000099397
|
or105-1
|
odorant receptor, family C, subfamily 105, member 1 |
| chr6_-_39051319 | 0.25 |
ENSDART00000155093
|
tns2b
|
tensin 2b |
| chr17_-_50355512 | 0.25 |
ENSDART00000041685
|
otofb
|
otoferlin b |
| chr5_+_7564644 | 0.25 |
ENSDART00000192173
|
CABZ01039096.1
|
|
| chr16_-_22729119 | 0.25 |
ENSDART00000132944
|
tlr19
|
toll-like receptor 19 |
| chr4_-_62503772 | 0.25 |
ENSDART00000108891
|
si:dkey-165b20.1
|
si:dkey-165b20.1 |
| chr8_+_23194212 | 0.24 |
ENSDART00000028946
ENSDART00000186842 ENSDART00000137815 ENSDART00000136914 ENSDART00000144855 ENSDART00000182689 |
tpd52l2a
|
tumor protein D52-like 2a |
| chr22_+_8489400 | 0.24 |
ENSDART00000129119
|
CABZ01046407.1
|
|
| chr18_+_17611627 | 0.24 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr16_+_30961822 | 0.24 |
ENSDART00000059187
|
gstk2
|
glutathione S-transferase kappa 2 |
| chr7_-_6100716 | 0.24 |
ENSDART00000173003
|
si:cabz01036006.1
|
si:cabz01036006.1 |
| chr2_-_47904043 | 0.23 |
ENSDART00000185328
ENSDART00000126740 |
ftr22
|
finTRIM family, member 22 |
| chr11_-_27874116 | 0.23 |
ENSDART00000180579
|
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr24_+_29912509 | 0.23 |
ENSDART00000168422
|
frrs1b
|
ferric-chelate reductase 1b |
| chr4_-_49580812 | 0.23 |
ENSDART00000180306
|
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr17_-_23609210 | 0.22 |
ENSDART00000064003
|
ifit15
|
interferon-induced protein with tetratricopeptide repeats 15 |
| chr2_+_11670270 | 0.22 |
ENSDART00000100524
|
ftr01
|
finTRIM family, member 1 |
| chr2_+_42318012 | 0.22 |
ENSDART00000138137
|
ftr08
|
finTRIM family, member 8 |
| chr20_+_46897504 | 0.22 |
ENSDART00000158124
|
si:ch73-21k16.1
|
si:ch73-21k16.1 |
| chr2_-_58902477 | 0.22 |
ENSDART00000061638
ENSDART00000172822 |
mau2
|
MAU2 sister chromatid cohesion factor |
| chr4_+_16787488 | 0.22 |
ENSDART00000143006
|
golt1ba
|
golgi transport 1Ba |
| chr2_+_47927026 | 0.21 |
ENSDART00000143023
|
ftr25
|
finTRIM family, member 25 |
| chr4_-_64141714 | 0.21 |
ENSDART00000128628
|
BX914205.3
|
|
| chr11_+_12175162 | 0.21 |
ENSDART00000125446
|
si:ch211-156l18.7
|
si:ch211-156l18.7 |
| chr17_+_12265095 | 0.21 |
ENSDART00000153812
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr5_+_4806851 | 0.20 |
ENSDART00000067599
|
angptl2a
|
angiopoietin-like 2a |
| chr11_+_30306606 | 0.20 |
ENSDART00000128276
ENSDART00000190222 |
ugt1b4
|
UDP glucuronosyltransferase 1 family, polypeptide B4 |
| chr23_+_44633858 | 0.20 |
ENSDART00000180728
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr19_+_11256617 | 0.20 |
ENSDART00000159577
|
si:ch73-171o20.1
|
si:ch73-171o20.1 |
| chr4_-_76488581 | 0.20 |
ENSDART00000174291
|
ftr51
|
finTRIM family, member 51 |
| chr18_-_25402715 | 0.19 |
ENSDART00000179852
|
gnrhr4
|
gonadotropin releasing hormone receptor 4 |
| chr5_+_24305877 | 0.19 |
ENSDART00000144226
|
ctsll
|
cathepsin L, like |
| chr12_-_20795867 | 0.18 |
ENSDART00000152835
ENSDART00000153424 |
nme2a
|
NME/NM23 nucleoside diphosphate kinase 2a |
| chr7_-_16214889 | 0.18 |
ENSDART00000076318
ENSDART00000173946 |
btr06
|
bloodthirsty-related gene family, member 6 |
| chr2_-_59285407 | 0.18 |
ENSDART00000181616
|
ftr34
|
finTRIM family, member 34 |
| chr17_-_45386823 | 0.18 |
ENSDART00000156002
|
tmem206
|
transmembrane protein 206 |
| chr23_+_36052944 | 0.18 |
ENSDART00000103149
|
hoxc13a
|
homeobox C13a |
| chr21_+_25533908 | 0.18 |
ENSDART00000185225
|
nlrc3l1
|
NLR family, CARD domain containing 3-like 1 |
| chr2_+_19578446 | 0.17 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr20_+_25711718 | 0.17 |
ENSDART00000033436
|
cep135
|
centrosomal protein 135 |
Network of associatons between targets according to the STRING database.
First level regulatory network of smad3a+smad3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0098581 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.3 | 0.9 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.3 | 4.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 0.8 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 1.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 1.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 1.5 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 0.7 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 1.3 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.6 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.3 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.4 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.2 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.1 | 1.3 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.1 | 0.2 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 0.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.6 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 1.0 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.4 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 1.3 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.2 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.5 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 0.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.3 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.9 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 2.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.4 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.4 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.6 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 2.5 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.3 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 1.1 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 4.2 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 0.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.1 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 1.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.6 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 2.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.4 | 1.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 0.8 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.2 | 1.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 1.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.6 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 0.3 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 4.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.7 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.4 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 1.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.7 | GO:0022884 | macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 1.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 1.3 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.6 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 1.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 2.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |