Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for smad2_smad10a+smad4a_smad1
Z-value: 1.50
Transcription factors associated with smad2_smad10a+smad4a_smad1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
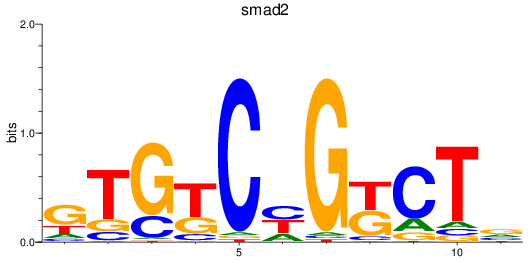
smad2
|
ENSDARG00000006389 | SMAD family member 2 |
|
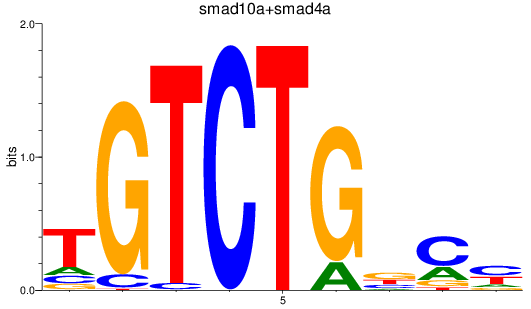
smad10a
|
ENSDARG00000045094 | si_dkey-222b8.1 |
|
smad10a
|
ENSDARG00000070428 | si_dkey-222b8.1 |
|
smad4a
|
ENSDARG00000075226 | SMAD family member 4a |
|
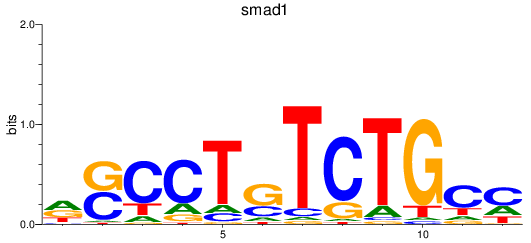
smad1
|
ENSDARG00000027199 | SMAD family member 1 |
|
smad1
|
ENSDARG00000112617 | SMAD family member 1 |
|
smad1
|
ENSDARG00000115674 | SMAD family member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:dkey-239n17.4 | dr11_v1_chr16_-_28878080_28878080 | 0.88 | 4.7e-02 | Click! |
| smad1 | dr11_v1_chr1_-_35929143_35929143 | 0.64 | 2.5e-01 | Click! |
| smad4a | dr11_v1_chr10_+_585719_585719 | 0.41 | 4.9e-01 | Click! |
| smad2 | dr11_v1_chr10_-_14943281_14943281 | 0.16 | 8.0e-01 | Click! |
| si:dkey-222b8.1 | dr11_v1_chr19_+_23932259_23932259 | -0.02 | 9.8e-01 | Click! |
Activity profile of smad2_smad10a+smad4a_smad1 motif
Sorted Z-values of smad2_smad10a+smad4a_smad1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_4932619 | 1.20 |
ENSDART00000103293
|
ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr24_-_28243186 | 1.19 |
ENSDART00000105691
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr20_-_7648325 | 1.04 |
ENSDART00000186541
|
CR848049.1
|
|
| chr23_-_39849155 | 0.90 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr4_-_16412084 | 0.77 |
ENSDART00000188460
|
dcn
|
decorin |
| chr19_-_41069573 | 0.68 |
ENSDART00000111982
ENSDART00000193142 |
sgce
|
sarcoglycan, epsilon |
| chr21_-_43616586 | 0.66 |
ENSDART00000036537
|
htr2cl2
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled-like 2 |
| chr13_+_31648271 | 0.64 |
ENSDART00000006648
|
mnat1
|
MNAT CDK-activating kinase assembly factor 1 |
| chr23_+_36460239 | 0.64 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr13_+_25433774 | 0.63 |
ENSDART00000141255
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr5_+_64900223 | 0.61 |
ENSDART00000191677
|
ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr19_-_5380770 | 0.55 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr10_-_10018794 | 0.55 |
ENSDART00000130734
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr19_+_42219165 | 0.50 |
ENSDART00000163192
|
CU896644.1
|
|
| chr4_-_21466825 | 0.50 |
ENSDART00000066897
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr11_-_13152524 | 0.48 |
ENSDART00000181440
ENSDART00000191997 |
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr10_+_20364009 | 0.48 |
ENSDART00000186139
ENSDART00000080395 |
golga7
|
golgin A7 |
| chr11_-_497854 | 0.46 |
ENSDART00000104520
|
cnbpb
|
CCHC-type zinc finger, nucleic acid binding protein b |
| chr11_+_45255774 | 0.45 |
ENSDART00000172838
|
aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr15_+_28303161 | 0.44 |
ENSDART00000087926
|
myo1cb
|
myosin Ic, paralog b |
| chr23_+_41831224 | 0.44 |
ENSDART00000171885
|
scp2b
|
sterol carrier protein 2b |
| chr23_-_24263474 | 0.43 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr3_+_26813058 | 0.39 |
ENSDART00000055537
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr18_+_49225552 | 0.39 |
ENSDART00000135026
|
si:ch211-136a13.1
|
si:ch211-136a13.1 |
| chr8_-_32497815 | 0.39 |
ENSDART00000122359
|
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr9_-_35875051 | 0.38 |
ENSDART00000013432
|
dnajc28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr8_-_32497581 | 0.37 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr11_+_40032790 | 0.36 |
ENSDART00000158809
|
si:dkey-264d12.1
|
si:dkey-264d12.1 |
| chr25_-_31433512 | 0.35 |
ENSDART00000067028
|
zgc:172145
|
zgc:172145 |
| chr14_-_237130 | 0.34 |
ENSDART00000164988
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr22_+_35068046 | 0.32 |
ENSDART00000161660
ENSDART00000169573 |
si:ch73-173h19.3
|
si:ch73-173h19.3 |
| chr7_-_18508815 | 0.31 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr7_+_9290929 | 0.30 |
ENSDART00000128530
|
snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr18_+_5549672 | 0.29 |
ENSDART00000184970
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr7_+_22767678 | 0.29 |
ENSDART00000137203
|
ponzr6
|
plac8 onzin related protein 6 |
| chr12_-_4070058 | 0.28 |
ENSDART00000042200
|
aldoab
|
aldolase a, fructose-bisphosphate, b |
| chr15_+_11427620 | 0.28 |
ENSDART00000168688
|
ndufc2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 |
| chr11_+_77526 | 0.27 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr13_+_33703784 | 0.26 |
ENSDART00000173361
|
macrod2
|
MACRO domain containing 2 |
| chr9_+_45605410 | 0.26 |
ENSDART00000136444
ENSDART00000007189 ENSDART00000158713 ENSDART00000182671 |
traf3ip1
|
TNF receptor-associated factor 3 interacting protein 1 |
| chr18_+_25003207 | 0.25 |
ENSDART00000099476
ENSDART00000132285 |
fam174b
|
family with sequence similarity 174, member B |
| chr24_-_4768955 | 0.24 |
ENSDART00000066834
|
agtr1b
|
angiotensin II receptor, type 1b |
| chr4_-_797831 | 0.24 |
ENSDART00000158970
ENSDART00000170012 |
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
| chr2_-_42628028 | 0.24 |
ENSDART00000179866
|
myo10
|
myosin X |
| chr19_-_43819582 | 0.23 |
ENSDART00000160879
ENSDART00000075902 |
klhl43
|
kelch-like family member 43 |
| chr2_+_54755172 | 0.23 |
ENSDART00000097864
|
ankrd12
|
ankyrin repeat domain 12 |
| chr10_-_10018120 | 0.22 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr12_+_15165736 | 0.22 |
ENSDART00000180398
|
adprh
|
ADP-ribosylarginine hydrolase |
| chr22_+_30446987 | 0.22 |
ENSDART00000146471
|
si:dkey-169i5.4
|
si:dkey-169i5.4 |
| chr14_-_25949951 | 0.22 |
ENSDART00000141304
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr14_+_45608576 | 0.22 |
ENSDART00000173248
|
si:ch211-276i12.11
|
si:ch211-276i12.11 |
| chr19_-_11425542 | 0.22 |
ENSDART00000177875
ENSDART00000080762 |
sept7a
|
septin 7a |
| chr3_-_56569968 | 0.21 |
ENSDART00000165317
|
SMIM10
|
Danio rerio uncharacterized LOC797202 (LOC797202), mRNA. |
| chr14_-_736575 | 0.21 |
ENSDART00000168611
|
tlr1
|
toll-like receptor 1 |
| chr15_+_37545855 | 0.21 |
ENSDART00000099456
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr3_-_21242460 | 0.20 |
ENSDART00000007293
|
tcap
|
titin-cap (telethonin) |
| chr20_+_23625387 | 0.20 |
ENSDART00000147945
ENSDART00000150497 |
palld
|
palladin, cytoskeletal associated protein |
| chr17_-_14671098 | 0.20 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr8_+_30742898 | 0.20 |
ENSDART00000018475
|
snrpd3
|
small nuclear ribonucleoprotein D3 polypeptide |
| chr25_+_32496877 | 0.19 |
ENSDART00000132698
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr9_-_21904695 | 0.19 |
ENSDART00000134768
|
lmo7a
|
LIM domain 7a |
| chr6_+_8172227 | 0.18 |
ENSDART00000146106
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr5_+_24287927 | 0.18 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr10_-_34976812 | 0.18 |
ENSDART00000170625
|
smad9
|
SMAD family member 9 |
| chr9_-_21936841 | 0.18 |
ENSDART00000144843
|
lmo7a
|
LIM domain 7a |
| chr21_-_13751535 | 0.18 |
ENSDART00000111666
|
npdc1a
|
neural proliferation, differentiation and control, 1a |
| chr23_+_40908583 | 0.18 |
ENSDART00000180933
|
LO017845.1
|
|
| chr5_+_18047111 | 0.18 |
ENSDART00000132164
|
hira
|
histone cell cycle regulator a |
| chr16_+_31542645 | 0.18 |
ENSDART00000163724
|
SLA (1 of many)
|
Src like adaptor |
| chr3_-_36846496 | 0.17 |
ENSDART00000055237
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr17_+_33433576 | 0.17 |
ENSDART00000077581
|
snap23.2
|
synaptosomal-associated protein 23.2 |
| chr7_-_72269049 | 0.17 |
ENSDART00000161497
|
CU463038.2
|
|
| chr25_+_32496723 | 0.17 |
ENSDART00000087978
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr13_+_23157053 | 0.17 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr18_+_50461981 | 0.17 |
ENSDART00000158761
|
CU896640.1
|
|
| chr19_-_3574060 | 0.17 |
ENSDART00000105120
|
tmem170b
|
transmembrane protein 170B |
| chr2_+_26240339 | 0.17 |
ENSDART00000191006
|
palm1b
|
paralemmin 1b |
| chr24_-_37272116 | 0.16 |
ENSDART00000022999
|
ube2ib
|
ubiquitin-conjugating enzyme E2Ib |
| chr17_+_50524573 | 0.16 |
ENSDART00000187974
|
CR382385.1
|
|
| chr23_+_19790962 | 0.16 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr24_-_28245872 | 0.16 |
ENSDART00000167861
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr13_+_45980163 | 0.16 |
ENSDART00000074547
ENSDART00000005195 |
bsdc1
|
BSD domain containing 1 |
| chr7_-_26601307 | 0.16 |
ENSDART00000188934
|
plscr3b
|
phospholipid scramblase 3b |
| chr9_+_23900703 | 0.16 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr23_+_45734011 | 0.16 |
ENSDART00000062415
|
CABZ01088025.1
|
|
| chr6_-_33925381 | 0.16 |
ENSDART00000137268
ENSDART00000145019 ENSDART00000141822 |
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr9_-_21230999 | 0.16 |
ENSDART00000150160
ENSDART00000102147 |
pla1a
|
phospholipase A1 member A |
| chr19_-_5369486 | 0.16 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr3_-_29928695 | 0.15 |
ENSDART00000151275
ENSDART00000151083 |
grna
|
granulin a |
| chr19_+_20177208 | 0.15 |
ENSDART00000166807
|
tra2a
|
transformer 2 alpha homolog |
| chr6_-_15653494 | 0.15 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr4_+_76957765 | 0.15 |
ENSDART00000129607
|
si:dkey-240n22.6
|
si:dkey-240n22.6 |
| chr1_+_59073203 | 0.15 |
ENSDART00000149937
ENSDART00000162201 |
MFAP4 (1 of many)
zgc:173915
|
si:zfos-2330d3.3 zgc:173915 |
| chr13_+_281214 | 0.15 |
ENSDART00000137572
|
mpc1
|
mitochondrial pyruvate carrier 1 |
| chr19_+_46113828 | 0.15 |
ENSDART00000159331
ENSDART00000161826 |
rbm24a
|
RNA binding motif protein 24a |
| chr21_-_17603182 | 0.15 |
ENSDART00000020048
ENSDART00000177270 |
gsna
|
gelsolin a |
| chr5_-_25583125 | 0.15 |
ENSDART00000031665
ENSDART00000145353 |
anxa1a
|
annexin A1a |
| chr6_-_29195642 | 0.15 |
ENSDART00000078625
|
dpt
|
dermatopontin |
| chr19_+_4061699 | 0.15 |
ENSDART00000158309
ENSDART00000166512 |
btr25
btr26
|
bloodthirsty-related gene family, member 25 bloodthirsty-related gene family, member 26 |
| chr13_+_22295905 | 0.14 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr15_+_28106498 | 0.14 |
ENSDART00000041707
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr2_-_59265521 | 0.14 |
ENSDART00000146341
ENSDART00000097799 |
ftr33
|
finTRIM family, member 33 |
| chr10_-_24759616 | 0.14 |
ENSDART00000079528
|
ilk
|
integrin-linked kinase |
| chr20_+_35085224 | 0.14 |
ENSDART00000040456
|
cdc42bpab
|
CDC42 binding protein kinase alpha (DMPK-like) b |
| chr8_+_554531 | 0.14 |
ENSDART00000193623
|
FO704758.2
|
|
| chr18_-_14337065 | 0.14 |
ENSDART00000135703
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr5_+_3892551 | 0.14 |
ENSDART00000134396
|
rpain
|
RPA interacting protein |
| chr6_-_52757276 | 0.14 |
ENSDART00000159154
|
AL954861.2
|
|
| chr4_-_76488581 | 0.14 |
ENSDART00000174291
|
ftr51
|
finTRIM family, member 51 |
| chr16_-_13595027 | 0.14 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr6_-_35439406 | 0.14 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr16_+_40340222 | 0.14 |
ENSDART00000190631
|
mettl6
|
methyltransferase like 6 |
| chr25_-_27819838 | 0.14 |
ENSDART00000067106
|
lmod2a
|
leiomodin 2 (cardiac) a |
| chr4_+_70351760 | 0.13 |
ENSDART00000159832
|
si:dkey-190j3.6
|
si:dkey-190j3.6 |
| chr14_+_6546516 | 0.13 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr14_-_12307522 | 0.13 |
ENSDART00000163900
|
myot
|
myotilin |
| chr22_-_11136625 | 0.13 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr19_-_43757568 | 0.13 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr14_+_51098036 | 0.13 |
ENSDART00000184118
|
CABZ01078594.1
|
|
| chr20_-_7293837 | 0.13 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr4_+_5156117 | 0.13 |
ENSDART00000067392
|
tigarb
|
tp53-induced glycolysis and apoptosis regulator b |
| chr11_-_3646793 | 0.13 |
ENSDART00000077344
|
itih1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr16_+_40301056 | 0.13 |
ENSDART00000058578
|
rspo3
|
R-spondin 3 |
| chr20_+_26349002 | 0.13 |
ENSDART00000152842
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr10_+_5268054 | 0.13 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr25_-_3892686 | 0.13 |
ENSDART00000043172
|
tmem258
|
transmembrane protein 258 |
| chr24_+_6353394 | 0.13 |
ENSDART00000165118
|
CR352329.1
|
|
| chr6_-_55423220 | 0.13 |
ENSDART00000158929
|
ctsa
|
cathepsin A |
| chr19_+_4068134 | 0.12 |
ENSDART00000158285
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr8_-_26388090 | 0.12 |
ENSDART00000147912
|
si:dkey-20d21.12
|
si:dkey-20d21.12 |
| chr6_-_42049350 | 0.12 |
ENSDART00000022949
|
parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr8_-_2230128 | 0.12 |
ENSDART00000140427
|
si:dkeyp-117b11.2
|
si:dkeyp-117b11.2 |
| chr13_+_22480496 | 0.12 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr23_-_874418 | 0.12 |
ENSDART00000133906
|
rbm10
|
RNA binding motif protein 10 |
| chr17_-_52091999 | 0.12 |
ENSDART00000019766
|
tgfb3
|
transforming growth factor, beta 3 |
| chr13_+_23176330 | 0.12 |
ENSDART00000168351
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr20_-_15922986 | 0.11 |
ENSDART00000189421
|
fam20b
|
family with sequence similarity 20, member B (H. sapiens) |
| chr17_+_53311618 | 0.11 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr12_+_48581588 | 0.11 |
ENSDART00000114996
|
CABZ01078415.1
|
|
| chr24_-_12958668 | 0.11 |
ENSDART00000178982
|
FITM1 (1 of many)
|
Danio rerio fat storage inducing transmembrane protein 1 (LOC792443), mRNA. |
| chr4_+_11458078 | 0.11 |
ENSDART00000037600
|
ankrd16
|
ankyrin repeat domain 16 |
| chr11_-_3647014 | 0.11 |
ENSDART00000171786
|
itih1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr15_-_47468085 | 0.11 |
ENSDART00000164438
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr11_+_141504 | 0.11 |
ENSDART00000086166
|
NCKAP1L
|
zgc:172352 |
| chr4_+_41602 | 0.11 |
ENSDART00000159640
|
phtf2
|
putative homeodomain transcription factor 2 |
| chr7_-_25895189 | 0.11 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr22_-_506522 | 0.11 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr25_+_37340722 | 0.11 |
ENSDART00000137025
|
pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr6_+_49881864 | 0.11 |
ENSDART00000075040
|
tubb1
|
tubulin, beta 1 class VI |
| chr9_-_34191627 | 0.11 |
ENSDART00000142664
|
dcaf6
|
ddb1 and cul4 associated factor 6 |
| chr1_-_9104631 | 0.11 |
ENSDART00000146642
|
si:ch211-14k19.8
|
si:ch211-14k19.8 |
| chr14_+_2243 | 0.11 |
ENSDART00000191193
|
CYTL1
|
cytokine like 1 |
| chr18_-_49020066 | 0.11 |
ENSDART00000174394
|
BX663503.3
|
|
| chr6_-_24103666 | 0.11 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr24_-_28306484 | 0.11 |
ENSDART00000148618
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr3_-_1434135 | 0.11 |
ENSDART00000149622
|
mgp
|
matrix Gla protein |
| chr13_+_22480857 | 0.11 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr5_+_36661058 | 0.11 |
ENSDART00000125653
|
capns1a
|
calpain, small subunit 1 a |
| chr6_+_60036767 | 0.10 |
ENSDART00000155009
|
tgm8
|
transglutaminase 8 |
| chr17_+_23968214 | 0.10 |
ENSDART00000183053
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr11_-_44647286 | 0.10 |
ENSDART00000169329
ENSDART00000158939 |
tomm20b
|
translocase of outer mitochondrial membrane 20b |
| chr3_+_28594713 | 0.10 |
ENSDART00000179799
|
sept12
|
septin 12 |
| chr19_-_7144548 | 0.10 |
ENSDART00000147177
ENSDART00000134850 |
psmb8a
psmb13a
|
proteasome subunit beta 8A proteasome subunit beta 13a |
| chr14_-_29799993 | 0.10 |
ENSDART00000133775
ENSDART00000005568 |
pdlim3b
|
PDZ and LIM domain 3b |
| chr2_-_42128714 | 0.10 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr12_+_8822717 | 0.10 |
ENSDART00000021628
|
reep3b
|
receptor accessory protein 3b |
| chr9_-_23922011 | 0.10 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr24_+_41610068 | 0.10 |
ENSDART00000159507
|
CABZ01044108.1
|
|
| chr18_-_5509616 | 0.10 |
ENSDART00000142945
|
bloc1s6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr8_+_22359881 | 0.10 |
ENSDART00000187867
|
zgc:153631
|
zgc:153631 |
| chr10_-_43334914 | 0.10 |
ENSDART00000146660
ENSDART00000190288 |
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr6_+_41463786 | 0.10 |
ENSDART00000065006
|
twf2a
|
twinfilin actin-binding protein 2a |
| chr16_-_39859119 | 0.10 |
ENSDART00000138388
|
rbms3
|
RNA binding motif, single stranded interacting protein |
| chr19_-_32487469 | 0.10 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr13_+_42124566 | 0.10 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr9_+_24095677 | 0.10 |
ENSDART00000150443
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr2_-_43135700 | 0.10 |
ENSDART00000098284
|
ftr14
|
finTRIM family, member 14 |
| chr14_+_9421510 | 0.10 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr14_-_34700633 | 0.10 |
ENSDART00000150358
|
ablim3
|
actin binding LIM protein family, member 3 |
| chr22_-_30935510 | 0.10 |
ENSDART00000133335
|
si:dkey-49n23.1
|
si:dkey-49n23.1 |
| chr20_+_34151670 | 0.10 |
ENSDART00000152870
ENSDART00000010329 ENSDART00000145852 |
arpc5b
|
actin related protein 2/3 complex, subunit 5B |
| chr15_-_419597 | 0.10 |
ENSDART00000058993
|
psph
|
phosphoserine phosphatase |
| chr12_-_628024 | 0.10 |
ENSDART00000158563
ENSDART00000152612 |
wu:fj29h11
si:ch73-301j1.1
|
wu:fj29h11 si:ch73-301j1.1 |
| chr12_-_9132682 | 0.10 |
ENSDART00000066471
|
adam8b
|
ADAM metallopeptidase domain 8b |
| chr18_-_44129151 | 0.10 |
ENSDART00000087339
|
cdon
|
cell adhesion associated, oncogene regulated |
| chr23_-_27479558 | 0.09 |
ENSDART00000013563
|
atf7a
|
activating transcription factor 7a |
| chr23_+_25893020 | 0.09 |
ENSDART00000144769
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr5_+_8964926 | 0.09 |
ENSDART00000091397
ENSDART00000164535 |
tnksb
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase b |
| chr4_+_50401133 | 0.09 |
ENSDART00000150644
|
si:dkey-156k2.7
|
si:dkey-156k2.7 |
| chr19_-_38611814 | 0.09 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr12_-_5505205 | 0.09 |
ENSDART00000092319
|
abi3b
|
ABI family, member 3b |
| chr19_-_24218942 | 0.09 |
ENSDART00000189198
|
BX547993.2
|
|
| chr20_+_54356540 | 0.09 |
ENSDART00000143591
|
znf410
|
zinc finger protein 410 |
| chr19_-_12193622 | 0.09 |
ENSDART00000041960
|
ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr5_-_26466169 | 0.09 |
ENSDART00000144035
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr18_+_8912113 | 0.09 |
ENSDART00000147467
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr2_-_43151358 | 0.09 |
ENSDART00000061170
ENSDART00000056161 |
ftr15
|
finTRIM family, member 15 |
| chr22_+_81799 | 0.09 |
ENSDART00000163661
|
zgc:112334
|
zgc:112334 |
| chr21_-_45728069 | 0.09 |
ENSDART00000185101
|
CU302316.1
|
|
| chr20_-_15922210 | 0.09 |
ENSDART00000152412
ENSDART00000152354 ENSDART00000152828 ENSDART00000013453 ENSDART00000152357 |
fam20b
|
family with sequence similarity 20, member B (H. sapiens) |
Network of associatons between targets according to the STRING database.
First level regulatory network of smad2_smad10a+smad4a_smad1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.2 | 0.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 1.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.5 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.4 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.1 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.4 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.4 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.1 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.2 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.4 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.3 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) cerebrospinal fluid circulation(GO:0090660) |
| 0.0 | 0.6 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0090024 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 0.8 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 1.5 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0045064 | T-helper 2 cell differentiation(GO:0045064) |
| 0.0 | 0.1 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.0 | 0.1 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.5 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0032206 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.2 | GO:0034205 | Notch receptor processing(GO:0007220) beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.7 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.2 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.0 | 0.1 | GO:0010660 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.0 | 0.1 | GO:0033632 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.1 | GO:0061614 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.2 | GO:2000403 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.2 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.0 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.0 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.0 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.0 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.0 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 0.0 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.6 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 1.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.0 | GO:0008352 | katanin complex(GO:0008352) |
| 0.0 | 0.1 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.0 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.2 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.3 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 1.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.2 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.3 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 1.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:1990825 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 1.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.5 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.0 | 1.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.0 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.0 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.0 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.0 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.6 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.0 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |