Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
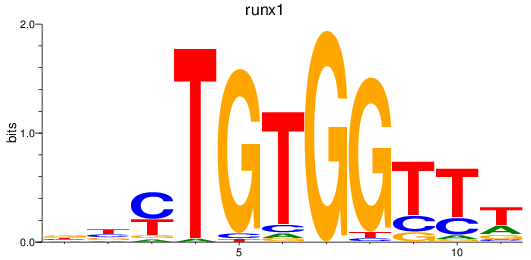
Results for runx1
Z-value: 2.66
Transcription factors associated with runx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
runx1
|
ENSDARG00000087646 | RUNX family transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| runx1 | dr11_v1_chr1_-_1402303_1402303 | -0.71 | 1.8e-01 | Click! |
Activity profile of runx1 motif
Sorted Z-values of runx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_5543677 | 2.70 |
ENSDART00000146161
ENSDART00000136189 |
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr17_-_2039511 | 2.30 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr8_-_21372446 | 2.28 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr8_+_21353878 | 2.01 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr7_-_7810348 | 1.94 |
ENSDART00000171984
|
cxcl19
|
chemokine (C-X-C motif) ligand 19 |
| chr4_-_77557279 | 1.92 |
ENSDART00000180113
|
AL935186.10
|
|
| chr7_+_31879986 | 1.87 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr24_-_9989634 | 1.86 |
ENSDART00000115275
|
zgc:152652
|
zgc:152652 |
| chr20_-_14054083 | 1.70 |
ENSDART00000009549
|
rhag
|
Rh associated glycoprotein |
| chr22_+_34616151 | 1.47 |
ENSDART00000155399
ENSDART00000104705 |
si:ch1073-214b20.2
|
si:ch1073-214b20.2 |
| chr4_+_77973876 | 1.43 |
ENSDART00000057423
|
terfa
|
telomeric repeat binding factor a |
| chr4_-_68563862 | 1.43 |
ENSDART00000182970
|
BX548011.2
|
|
| chr14_+_38786298 | 1.42 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr7_+_31879649 | 1.40 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr9_+_23770666 | 1.38 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr7_+_39706004 | 1.37 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr4_+_5537101 | 1.36 |
ENSDART00000008692
|
si:dkey-14d8.7
|
si:dkey-14d8.7 |
| chr10_-_15128771 | 1.27 |
ENSDART00000101261
|
spp1
|
secreted phosphoprotein 1 |
| chr3_-_16784280 | 1.23 |
ENSDART00000137108
ENSDART00000137276 |
si:dkey-30j10.5
|
si:dkey-30j10.5 |
| chr8_+_47099033 | 1.14 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr6_-_43449013 | 1.12 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr9_+_30464641 | 1.12 |
ENSDART00000128357
|
gja5a
|
gap junction protein, alpha 5a |
| chr19_-_7144548 | 1.10 |
ENSDART00000147177
ENSDART00000134850 |
psmb8a
psmb13a
|
proteasome subunit beta 8A proteasome subunit beta 13a |
| chr2_+_36898982 | 1.07 |
ENSDART00000084859
|
rabgap1l2
|
RAB GTPase activating protein 1-like 2 |
| chr12_+_34273240 | 1.06 |
ENSDART00000037904
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr22_+_21324398 | 1.05 |
ENSDART00000168509
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr22_+_19486711 | 1.05 |
ENSDART00000144389
ENSDART00000139256 ENSDART00000160855 |
si:dkey-78l4.10
|
si:dkey-78l4.10 |
| chr19_+_4068134 | 1.04 |
ENSDART00000158285
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr17_-_8899323 | 1.04 |
ENSDART00000081590
|
nkl.1
|
NK-lysin tandem duplicate 1 |
| chr19_-_40191358 | 1.03 |
ENSDART00000183919
|
grn1
|
granulin 1 |
| chr16_-_17300030 | 1.03 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr1_-_38195012 | 1.02 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr5_+_57658898 | 1.02 |
ENSDART00000074268
ENSDART00000124568 |
zgc:153929
|
zgc:153929 |
| chr10_+_2742499 | 1.01 |
ENSDART00000122847
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr21_-_28439596 | 1.01 |
ENSDART00000089980
ENSDART00000132844 |
rasgrp2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr19_-_5372572 | 1.00 |
ENSDART00000151326
|
krt17
|
keratin 17 |
| chr5_+_13373593 | 1.00 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr18_+_35742838 | 1.00 |
ENSDART00000088504
ENSDART00000140386 |
rasgrp4
|
RAS guanyl releasing protein 4 |
| chr22_-_5323482 | 0.99 |
ENSDART00000145785
|
s1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr3_-_34022076 | 0.99 |
ENSDART00000150907
ENSDART00000151092 |
ighv1-4
ighv1-4
|
immunoglobulin heavy variable 1-4 immunoglobulin heavy variable 1-4 |
| chr1_+_55703120 | 0.98 |
ENSDART00000141089
|
adgre6
|
adhesion G protein-coupled receptor E6 |
| chr10_+_9550419 | 0.98 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr4_-_13502549 | 0.96 |
ENSDART00000140366
|
si:ch211-266a5.12
|
si:ch211-266a5.12 |
| chr21_-_43616586 | 0.94 |
ENSDART00000036537
|
htr2cl2
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled-like 2 |
| chr10_+_17088261 | 0.94 |
ENSDART00000132103
|
si:dkey-106l3.7
|
si:dkey-106l3.7 |
| chr3_-_21382491 | 0.93 |
ENSDART00000016562
|
itga2b
|
integrin, alpha 2b |
| chr14_+_22022441 | 0.92 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr23_-_36316352 | 0.92 |
ENSDART00000014840
|
nfe2
|
nuclear factor, erythroid 2 |
| chr17_+_5426087 | 0.92 |
ENSDART00000131430
ENSDART00000188636 |
runx2a
|
runt-related transcription factor 2a |
| chr15_+_37412883 | 0.91 |
ENSDART00000156474
|
zbtb32
|
zinc finger and BTB domain containing 32 |
| chr21_+_25198637 | 0.89 |
ENSDART00000164972
|
si:dkey-183i3.6
|
si:dkey-183i3.6 |
| chr13_-_25305179 | 0.89 |
ENSDART00000110064
|
plaua
|
plasminogen activator, urokinase a |
| chr4_-_13518381 | 0.88 |
ENSDART00000067153
|
ifng1-1
|
interferon, gamma 1-1 |
| chr17_+_1627379 | 0.88 |
ENSDART00000184050
|
LO018432.1
|
|
| chr8_+_25359394 | 0.88 |
ENSDART00000111248
|
si:dkey-183p4.10
|
si:dkey-183p4.10 |
| chr13_-_37122217 | 0.87 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr8_-_49207319 | 0.86 |
ENSDART00000022870
|
fam110a
|
family with sequence similarity 110, member A |
| chr21_+_11560153 | 0.86 |
ENSDART00000065842
|
cd8a
|
CD8a molecule |
| chr16_-_22194400 | 0.86 |
ENSDART00000186042
|
il6r
|
interleukin 6 receptor |
| chr22_+_19494996 | 0.85 |
ENSDART00000140829
ENSDART00000135168 |
si:dkey-78l4.10
|
si:dkey-78l4.10 |
| chr2_-_40199780 | 0.85 |
ENSDART00000113901
|
ccl34a.4
|
chemokine (C-C motif) ligand 34a, duplicate 4 |
| chr1_+_54650048 | 0.84 |
ENSDART00000141207
|
si:ch211-202h22.7
|
si:ch211-202h22.7 |
| chr25_-_13202208 | 0.84 |
ENSDART00000168155
|
si:ch211-194m7.4
|
si:ch211-194m7.4 |
| chr3_+_14641962 | 0.84 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr1_+_109798 | 0.83 |
ENSDART00000165402
|
F7 (1 of many)
|
zgc:163025 |
| chr14_-_11430566 | 0.83 |
ENSDART00000137154
ENSDART00000091158 |
irg1l
|
immunoresponsive gene 1, like |
| chr4_-_26108053 | 0.83 |
ENSDART00000066951
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr6_+_45932276 | 0.83 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr1_-_12027 | 0.83 |
ENSDART00000164359
|
rpl24
|
ribosomal protein L24 |
| chr9_+_48108174 | 0.83 |
ENSDART00000077260
|
cxcr2
|
chemokine (C-X-C motif) receptor 2 |
| chr24_-_25244637 | 0.82 |
ENSDART00000153798
|
hhla2b.2
|
HERV-H LTR-associating 2b, tandem duplicate 2 |
| chr23_+_17417539 | 0.81 |
ENSDART00000182605
|
BX649300.2
|
|
| chr2_+_51028269 | 0.81 |
ENSDART00000161254
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr2_-_5475910 | 0.81 |
ENSDART00000100954
ENSDART00000172143 ENSDART00000132496 |
proca
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr12_+_804314 | 0.81 |
ENSDART00000082169
|
wfikkn2b
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2b |
| chr8_+_25342896 | 0.80 |
ENSDART00000129032
|
CR847543.1
|
|
| chr6_-_13498745 | 0.80 |
ENSDART00000027684
ENSDART00000189438 |
mylkb
|
myosin light chain kinase b |
| chr25_+_19680962 | 0.80 |
ENSDART00000167260
|
si:dkeyp-110c12.3
|
si:dkeyp-110c12.3 |
| chr8_+_19356072 | 0.79 |
ENSDART00000063272
|
mpeg1.2
|
macrophage expressed 1, tandem duplicate 2 |
| chr7_-_60831082 | 0.78 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr16_-_51254694 | 0.77 |
ENSDART00000148894
|
serpinb14
|
serpin peptidase inhibitor, clade B (ovalbumin), member 14 |
| chr19_+_4062101 | 0.77 |
ENSDART00000166773
|
btr25
|
bloodthirsty-related gene family, member 25 |
| chr3_-_7948799 | 0.77 |
ENSDART00000163714
|
trim35-24
|
tripartite motif containing 35-24 |
| chr22_+_9772754 | 0.77 |
ENSDART00000130194
|
BX664625.2
|
|
| chr1_+_55140970 | 0.77 |
ENSDART00000039807
|
mb
|
myoglobin |
| chr11_+_37216668 | 0.76 |
ENSDART00000173076
|
zgc:112265
|
zgc:112265 |
| chr3_+_22335030 | 0.76 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr17_+_5976683 | 0.76 |
ENSDART00000110276
|
zgc:194275
|
zgc:194275 |
| chr16_-_17056630 | 0.76 |
ENSDART00000138715
|
plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr20_+_1398564 | 0.75 |
ENSDART00000002242
|
leg1.2
|
liver-enriched gene 1, tandem duplicate 2 |
| chr3_+_3598555 | 0.75 |
ENSDART00000191152
|
CR589947.3
|
|
| chr20_+_54034677 | 0.75 |
ENSDART00000173317
ENSDART00000173215 |
si:dkey-241l7.2
|
si:dkey-241l7.2 |
| chr5_+_43783360 | 0.75 |
ENSDART00000019431
|
nos2a
|
nitric oxide synthase 2a, inducible |
| chr7_+_24523017 | 0.75 |
ENSDART00000077047
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr10_-_8053385 | 0.74 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr3_+_14558188 | 0.74 |
ENSDART00000143904
|
epor
|
erythropoietin receptor |
| chr4_+_39766931 | 0.74 |
ENSDART00000138663
|
si:dkeyp-85d8.1
|
si:dkeyp-85d8.1 |
| chr18_+_22109379 | 0.74 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr6_-_15096556 | 0.73 |
ENSDART00000185327
|
fhl2b
|
four and a half LIM domains 2b |
| chr13_+_41819817 | 0.73 |
ENSDART00000185778
|
CABZ01066611.1
|
|
| chr19_-_35428815 | 0.73 |
ENSDART00000169006
ENSDART00000003167 |
ak2
|
adenylate kinase 2 |
| chr14_+_34501245 | 0.73 |
ENSDART00000131424
|
lcp2b
|
lymphocyte cytosolic protein 2b |
| chr16_-_31675669 | 0.72 |
ENSDART00000168848
ENSDART00000158331 |
c1r
|
complement component 1, r subcomponent |
| chr19_-_40192249 | 0.72 |
ENSDART00000051972
|
grn1
|
granulin 1 |
| chr12_+_38940731 | 0.72 |
ENSDART00000193360
|
si:dkey-239b22.1
|
si:dkey-239b22.1 |
| chr11_+_43431289 | 0.72 |
ENSDART00000192526
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr2_+_32796873 | 0.72 |
ENSDART00000077511
|
ccr9a
|
chemokine (C-C motif) receptor 9a |
| chr14_-_22118055 | 0.71 |
ENSDART00000179892
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr3_+_30968176 | 0.71 |
ENSDART00000186266
|
prf1.9
|
perforin 1.9 |
| chr22_-_31020690 | 0.71 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr22_+_9862243 | 0.71 |
ENSDART00000105942
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr1_-_7951002 | 0.71 |
ENSDART00000138187
|
si:dkey-79f11.8
|
si:dkey-79f11.8 |
| chr7_+_41295974 | 0.70 |
ENSDART00000173568
ENSDART00000173544 |
si:dkey-86l18.10
|
si:dkey-86l18.10 |
| chr16_-_31686602 | 0.70 |
ENSDART00000170357
|
c1s
|
complement component 1, s subcomponent |
| chr9_-_3496548 | 0.69 |
ENSDART00000102876
|
cybrd1
|
cytochrome b reductase 1 |
| chr3_+_19621034 | 0.69 |
ENSDART00000025358
|
itgb3a
|
integrin beta 3a |
| chr1_-_43933271 | 0.69 |
ENSDART00000121605
|
scpp6
|
secretory calcium-binding phosphoprotein 6 |
| chr25_-_23526058 | 0.69 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr22_-_10470663 | 0.69 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr1_-_53476758 | 0.69 |
ENSDART00000074221
|
b3gnt2a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2a |
| chr4_+_76735113 | 0.68 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr10_+_16756546 | 0.68 |
ENSDART00000168624
|
CABZ01024659.1
|
|
| chr8_+_37749263 | 0.68 |
ENSDART00000108556
ENSDART00000147942 |
npm2a
|
nucleophosmin/nucleoplasmin, 2a |
| chr22_-_30770751 | 0.68 |
ENSDART00000172115
|
AL831726.2
|
|
| chr13_-_33170733 | 0.68 |
ENSDART00000057382
|
fbln5
|
fibulin 5 |
| chr14_-_46198373 | 0.68 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr14_+_29941445 | 0.68 |
ENSDART00000181761
|
fam149a
|
family with sequence similarity 149 member A |
| chr4_+_37952218 | 0.67 |
ENSDART00000186865
|
si:dkeyp-82b4.2
|
si:dkeyp-82b4.2 |
| chr6_+_34868156 | 0.67 |
ENSDART00000149364
|
il23r
|
interleukin 23 receptor |
| chr15_-_33965440 | 0.67 |
ENSDART00000163841
|
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr13_-_865193 | 0.67 |
ENSDART00000187053
|
AL929536.7
|
|
| chr18_+_31073265 | 0.66 |
ENSDART00000023539
|
cyba
|
cytochrome b-245, alpha polypeptide |
| chr3_-_32958505 | 0.66 |
ENSDART00000147374
ENSDART00000136919 |
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr3_+_13569806 | 0.66 |
ENSDART00000170105
|
si:ch73-106n3.2
|
si:ch73-106n3.2 |
| chr5_+_69856153 | 0.66 |
ENSDART00000124128
ENSDART00000073663 |
ugt2a6
|
UDP glucuronosyltransferase 2 family, polypeptide A6 |
| chr2_-_16159203 | 0.66 |
ENSDART00000153480
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr11_-_30630628 | 0.65 |
ENSDART00000103265
|
zgc:158773
|
zgc:158773 |
| chr24_+_13735616 | 0.65 |
ENSDART00000184267
|
msc
|
musculin (activated B-cell factor-1) |
| chr6_-_7438584 | 0.65 |
ENSDART00000053776
|
fkbp11
|
FK506 binding protein 11 |
| chr1_+_57187794 | 0.65 |
ENSDART00000152485
|
si:dkey-27j5.9
|
si:dkey-27j5.9 |
| chr13_-_45155792 | 0.65 |
ENSDART00000163556
|
runx3
|
runt-related transcription factor 3 |
| chr3_-_22356568 | 0.65 |
ENSDART00000058507
|
ifnphi2
|
interferon phi 2 |
| chr6_-_27108844 | 0.65 |
ENSDART00000073883
|
dtymk
|
deoxythymidylate kinase (thymidylate kinase) |
| chr17_-_10838434 | 0.65 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr11_-_12198765 | 0.64 |
ENSDART00000104203
ENSDART00000128364 ENSDART00000166887 ENSDART00000041533 |
krt95
|
kertain 95 |
| chr10_-_17501528 | 0.64 |
ENSDART00000144847
|
slc2a11l
|
solute carrier family 2 (facilitated glucose transporter), member 11-like |
| chr2_-_37465517 | 0.64 |
ENSDART00000139983
|
si:dkey-57k2.6
|
si:dkey-57k2.6 |
| chr7_+_53156810 | 0.63 |
ENSDART00000189816
|
cdh29
|
cadherin 29 |
| chr4_-_13613148 | 0.63 |
ENSDART00000067164
ENSDART00000111247 |
irf5
|
interferon regulatory factor 5 |
| chr3_-_7990516 | 0.63 |
ENSDART00000167877
|
TRIM35 (1 of many)
|
si:ch211-175l6.2 |
| chr11_-_21304452 | 0.62 |
ENSDART00000163008
|
RASSF5
|
si:dkey-85p17.3 |
| chr13_-_20381485 | 0.62 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr7_+_35229805 | 0.62 |
ENSDART00000173911
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr15_-_38129845 | 0.62 |
ENSDART00000057095
|
si:dkey-24p1.1
|
si:dkey-24p1.1 |
| chr24_-_25098719 | 0.62 |
ENSDART00000193651
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr5_+_65991152 | 0.62 |
ENSDART00000097756
|
lcn15
|
lipocalin 15 |
| chr7_-_52417060 | 0.61 |
ENSDART00000148579
|
myzap
|
myocardial zonula adherens protein |
| chr16_+_1383914 | 0.61 |
ENSDART00000185089
|
cers2b
|
ceramide synthase 2b |
| chr3_-_79366 | 0.61 |
ENSDART00000114289
|
zgc:165518
|
zgc:165518 |
| chr3_-_29870848 | 0.61 |
ENSDART00000186457
|
rpl3
|
ribosomal protein L3 |
| chr4_-_12795436 | 0.61 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr9_+_50175366 | 0.61 |
ENSDART00000170352
|
cobll1b
|
cordon-bleu WH2 repeat protein-like 1b |
| chr19_+_3849378 | 0.60 |
ENSDART00000166218
ENSDART00000159228 |
oscp1a
|
organic solute carrier partner 1a |
| chr19_+_37118547 | 0.60 |
ENSDART00000103163
|
cx30.9
|
connexin 30.9 |
| chr23_-_43486714 | 0.60 |
ENSDART00000169726
|
e2f1
|
E2F transcription factor 1 |
| chr1_-_55008882 | 0.60 |
ENSDART00000083572
|
zgc:136864
|
zgc:136864 |
| chr2_-_37862380 | 0.60 |
ENSDART00000186005
|
si:ch211-284o19.8
|
si:ch211-284o19.8 |
| chr19_+_40350468 | 0.59 |
ENSDART00000087444
|
hepacam2
|
HEPACAM family member 2 |
| chr14_-_26498196 | 0.59 |
ENSDART00000054175
ENSDART00000145625 ENSDART00000183347 ENSDART00000191084 ENSDART00000191143 |
smad5
|
SMAD family member 5 |
| chr9_-_43082945 | 0.59 |
ENSDART00000142257
|
ccdc141
|
coiled-coil domain containing 141 |
| chr7_+_4694924 | 0.59 |
ENSDART00000144873
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr3_+_55114097 | 0.59 |
ENSDART00000121686
|
hbbe1.1
|
hemoglobin beta embryonic-1.1 |
| chr24_+_19518570 | 0.59 |
ENSDART00000056081
|
sulf1
|
sulfatase 1 |
| chr13_-_15982707 | 0.59 |
ENSDART00000186911
ENSDART00000181072 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr23_-_9925568 | 0.59 |
ENSDART00000081268
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr20_+_192170 | 0.59 |
ENSDART00000189675
|
cx28.8
|
connexin 28.8 |
| chr25_-_14637660 | 0.59 |
ENSDART00000143666
|
nav2b
|
neuron navigator 2b |
| chr8_+_6576940 | 0.58 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr2_-_16159491 | 0.58 |
ENSDART00000110059
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr24_+_38205798 | 0.58 |
ENSDART00000141532
|
igl3v1
|
immunoglobulin light 3 variable 1 |
| chr22_+_18156000 | 0.58 |
ENSDART00000143483
ENSDART00000136133 |
nr2c2ap
|
nuclear receptor 2C2-associated protein |
| chr6_-_7769178 | 0.58 |
ENSDART00000191701
ENSDART00000149823 |
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr22_+_6096303 | 0.58 |
ENSDART00000139276
|
zgc:171887
|
zgc:171887 |
| chr5_+_9360394 | 0.58 |
ENSDART00000124642
|
FP236810.2
|
|
| chr24_-_36680261 | 0.58 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr2_+_36620011 | 0.57 |
ENSDART00000177428
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr17_+_6382275 | 0.57 |
ENSDART00000056324
|
CR628323.2
|
|
| chr21_+_22985078 | 0.56 |
ENSDART00000156491
|
lpar6b
|
lysophosphatidic acid receptor 6b |
| chr9_+_9296222 | 0.56 |
ENSDART00000141325
|
si:ch211-214p13.8
|
si:ch211-214p13.8 |
| chr19_+_19786117 | 0.56 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr25_-_7974494 | 0.56 |
ENSDART00000171446
|
hal
|
histidine ammonia-lyase |
| chr17_-_36529016 | 0.56 |
ENSDART00000025019
|
colec11
|
collectin sub-family member 11 |
| chr18_+_27571448 | 0.56 |
ENSDART00000147886
|
cd82b
|
CD82 molecule b |
| chr16_-_30885838 | 0.55 |
ENSDART00000131356
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr23_+_10347851 | 0.55 |
ENSDART00000127667
|
krt18
|
keratin 18 |
| chr20_+_54027943 | 0.55 |
ENSDART00000153400
ENSDART00000152961 |
si:dkey-241l7.3
|
si:dkey-241l7.3 |
| chr22_-_10117918 | 0.55 |
ENSDART00000177475
ENSDART00000128590 |
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr19_+_32856907 | 0.55 |
ENSDART00000148232
|
rpl30
|
ribosomal protein L30 |
| chr4_-_26107841 | 0.54 |
ENSDART00000172012
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr25_+_4541211 | 0.54 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of runx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.7 | 3.3 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.4 | 1.7 | GO:0015840 | urea transport(GO:0015840) |
| 0.4 | 1.4 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.3 | 0.8 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.3 | 1.1 | GO:0009265 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) 2'-deoxyribonucleotide biosynthetic process(GO:0009265) deoxyribose phosphate biosynthetic process(GO:0046385) |
| 0.2 | 0.6 | GO:0019557 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.2 | 0.5 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.2 | 0.7 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.2 | 1.0 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.2 | 0.5 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 0.8 | GO:0030823 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 2.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 1.2 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 1.0 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 1.0 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.8 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.1 | 1.5 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.8 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.5 | GO:0050996 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) positive regulation of lipid catabolic process(GO:0050996) |
| 0.1 | 0.4 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.6 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.7 | GO:0000730 | DNA recombinase assembly(GO:0000730) |
| 0.1 | 0.4 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.1 | 0.7 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 0.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 0.2 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 9.3 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.1 | 0.6 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 0.3 | GO:0060148 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 0.4 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.1 | 0.7 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.5 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.6 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 0.3 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 0.3 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.1 | 0.9 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.1 | 1.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.3 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.5 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 | 0.4 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.4 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.4 | GO:1904105 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 0.3 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 1.6 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.9 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.1 | 0.2 | GO:0034398 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.2 | GO:0061213 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.6 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.4 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 1.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.2 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.2 | GO:0051099 | cytidine to uridine editing(GO:0016554) positive regulation of DNA binding(GO:0043388) positive regulation of binding(GO:0051099) |
| 0.1 | 0.5 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.8 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 1.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.9 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 0.3 | GO:0070199 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 0.6 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 1.0 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.3 | GO:0051661 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.3 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.3 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.4 | GO:1900117 | regulation of execution phase of apoptosis(GO:1900117) negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.2 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.4 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.1 | 0.6 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 0.4 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.2 | GO:0046833 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 0.2 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.1 | 0.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 0.3 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.2 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.6 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.3 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 | 0.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 0.7 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 0.6 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.1 | 0.4 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 1.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 2.9 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.7 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.8 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.4 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.4 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.5 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 5.9 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.4 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.4 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.6 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 2.2 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.3 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.2 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.1 | GO:0050968 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 3.7 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 1.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.2 | GO:0060420 | regulation of heart growth(GO:0060420) |
| 0.0 | 0.3 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.2 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.0 | 1.0 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 1.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 1.0 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.1 | GO:0033512 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.4 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.3 | GO:0070874 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.6 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.5 | GO:0006956 | complement activation(GO:0006956) protein activation cascade(GO:0072376) |
| 0.0 | 0.9 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 1.5 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.1 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.4 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 1.0 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 0.3 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.2 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.3 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) protein-lipid complex assembly(GO:0065005) |
| 0.0 | 0.1 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.7 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.2 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 0.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.2 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.4 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.6 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0009204 | deoxyribonucleoside triphosphate catabolic process(GO:0009204) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.5 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.1 | GO:0035739 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) negative regulation of alpha-beta T cell proliferation(GO:0046642) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.2 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.0 | 1.2 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.7 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 1.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.7 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 1.4 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 0.8 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.0 | 0.2 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.4 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.5 | GO:0002685 | regulation of leukocyte migration(GO:0002685) |
| 0.0 | 0.5 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.5 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:1903038 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 1.8 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.3 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.1 | GO:0097241 | cannabinoid signaling pathway(GO:0038171) hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.1 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.0 | 0.1 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.5 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.7 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.0 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
| 0.0 | 0.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.0 | GO:0032263 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.1 | GO:0016117 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.0 | 1.2 | GO:0002768 | immune response-regulating cell surface receptor signaling pathway(GO:0002768) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.0 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.0 | 0.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.6 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.2 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.6 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.0 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.0 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.4 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.3 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.5 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 0.1 | GO:0030325 | adrenal gland development(GO:0030325) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0033065 | Rad51C-XRCC3 complex(GO:0033065) |
| 0.2 | 0.7 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.2 | 1.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.5 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 0.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 1.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.5 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 1.7 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.3 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 0.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.3 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.3 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.3 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.6 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 0.5 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 0.6 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.3 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 1.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.5 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 1.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 2.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 6.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.8 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 2.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.0 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 19.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.0 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.1 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 2.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.0 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0045335 | phagocytic vesicle(GO:0045335) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.7 | 2.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.3 | 1.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.3 | 3.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 0.8 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 2.0 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 0.7 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.2 | 0.6 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 0.6 | GO:0042806 | fucose binding(GO:0042806) |
| 0.2 | 0.5 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.2 | 1.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 1.4 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.4 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.7 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.6 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 1.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.4 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.6 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.4 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 0.5 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.3 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.1 | 0.8 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.4 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.5 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 2.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 4.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.3 | GO:0032138 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.3 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 1.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.3 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 1.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.6 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 1.0 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.2 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.3 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.1 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.9 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.3 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 0.2 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.1 | 0.8 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.4 | GO:0019825 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.3 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.3 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 1.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.2 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 5.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.8 | GO:0022884 | macromolecule transmembrane transporter activity(GO:0022884) |
| 0.1 | 1.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.4 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.1 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.7 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 1.7 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 10.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.3 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.4 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0042165 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.0 | 0.3 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.5 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.4 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.5 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 1.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 1.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.4 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.2 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.1 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) |
| 0.0 | 1.0 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.5 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 1.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 1.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 1.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 1.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 3.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 6.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.0 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 1.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.4 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 1.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.0 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 1.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.7 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.6 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 3.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 1.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.9 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.9 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 1.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 1.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 3.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.8 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME DNA STRAND ELONGATION | Genes involved in DNA strand elongation |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |